Abstract
In developed countries, colorectal cancer (CRC) is the third most common malignancy, but it is the second most frequent cause of cancer-related death. Clinicians are still faced with numerous challenges in the treatment of this disease, and future approaches which target the molecular features of the disorder will be critical for success in this disease setting. Genetic analyses of many solid tumours have shown that up to 100 protein-encoding genes are mutated. Within CRC, numerous genetic alterations have been identified in a number of pathways. Therefore, understanding the molecular pathology of CRC may present information on potential routes for treatment and may also provide valuable prognostic information. This will be particularly pertinent for molecularly targeted treatments, such as anti-vascular endothelial growth factor therapies and anti-epidermal growth factor receptor (EGFR) monoclonal antibody therapy. KRAS and BRAF mutations have been shown to predict response to anti-EGFR therapy. As EGFR can also signal via the phosphatidylinositol 3-kinase (PI3K) kinase pathway, there is considerable interest in the potential roles of members of this pathway (such as PI3K and PTEN) in predicting treatment response. Therefore, a combined approach of new techniques that allow identification of these biomarkers alongside interdisciplinary approaches to the treatment of advanced CRC will aid in the treatment decision-making process and may also serve to guide future therapeutic approaches.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Review
Introduction to colorectal cancer
Colorectal cancer (CRC) is the fourth leading cause of cancer-related mortality worldwide, accounting for over 600,000 deaths annually [1]. Estimates for CRC in the 40 European countries studied in 2008 [1] indicated that CRC was the most common cancer (436,000 cases, 13.6% of total) and the second most common cause of death from cancer (212,000 deaths, 12.3% of total). Recent genome-wide analyses of solid tumours, including CRC, have shown mutations in between 20 and 100 protein-encoding genes [2–7]. A number of key genetic and epigenetic alterations which lead to malignant transformation have been identified in CRC, and these include aberrations in genes involved in the chromosomal instability (CIS) pathway, the microsatellite instability (MSI) pathway, the hMYH pathway and the CpG island methylation pathway [8]. Recent data have shown that KRAS and BRAF mutations predict response to anti-epidermal growth factor receptor (EGFR) therapy reviewed in [9]. Great strides have been made in the early detection and diagnosis of CRC, including population-based screening, which has the potential to prevent up to 60% of CRC deaths reviewed in [10]. Despite this, however, up to 56% of newly diagnosed CRC patients present with either nodal or distant metastases [11]. Prognosis is poor for these patients, with an overall 5-year survival rate of 6.6%–11.9% for Dukes D patients [11, 12]. Therefore, further developments are essential in order to increase the 5-year survival rate and to improve the overall quality of life (QoL) for patients with this disease.
The role of predictive, preventive and personalised medicine in CRC
Predictive, preventive and personalised medicine (PPPM) endeavours to promote a paradigm shift in our current healthcare approach. The PPPM approach aims to predict individual predisposition before onset of the disease, to provide targeted preventive measures and to create personalised treatment algorithms tailored to the individual. The concept aims to move from delayed intervention to predictive medicine tailored to the person, from reactive to preventive medicine and from disease to wellness. It is hoped that this will provide a more cost-effective management of major diseases, such as cancer, in the future. The critical role of PPPM in the modernisation of healthcare systems has been acknowledged as a priority by global and regional organisations and health-related institutions, such as the Organisation of United Nations, the European Union and The National Institutes of Health. In CRC, the potential value of biomarkers for PPPM is strong, and there have been recent increased efforts to incorporate the use of such markers into healthcare systems. Biomarkers could be used to:
-
Identify disease predisposition
-
Identify early disease and aid appropriate timely treatment intervention
-
Aid molecular classification of the disease, with a view to provide better disease understanding and more effective, targeted treatment options
-
Identify patient populations that are more likely to derive clinical benefit from current and future treatment options.
Whilst the identification of biomarkers of predisposition and of early stage disease is critical, the focus of this review article will be on the use of markers as an aid to the classification of CRC and their role as potential companion diagnostics.
Classification of CRC
Classification of CRC has traditionally been based on histopathological features. Molecular studies have allowed a significant appreciation of the heterogeneous nature of CRC. However, it has long been known, based on morphological criteria, that CRC is not a homogenous disease. For example, even before the advent of molecular classification of tumours, it was noted that the rare, but histologically distinct ‘medullary carcinoma’ occurs almost exclusively on the right side of the colon and is associated with an improved prognosis compared to the standard histological types [13].
More recently, the molecular changes underlying these phenotypical appearances have been elucidated. To use the example above, it has now been shown that medullary carcinoma of the colon shows MSI with loss of the DNA mismatch repair (MMR) enzymes, such as MLH-1, MSH-2, MSH-6 and PMS-2 [13]. It is also now known that this molecular signature is shared by the more common serrated tumour pathway. This loss of DNA mismatch repair activity may be caused by mutation of one of these genes, as seen in hereditary non-polyposis colorectal cancer (HNPCC). More commonly, these tumours arise sporadically, and the loss of MMR function is due to epigenetic silencing of one of the genes, as a result of promoter methylation. High-level MSI (MSI-H) comprises 15% of sporadic CRC, and these are positively correlated with patients being female, over 60 years of age, having BRAF mutations and being right-sided tumours [14]. It is interesting to note that although there is a continuous increase in the rate of MSI-H tumours as one progresses proximally from the rectum to the ascending colon, the previously popular view of dichotomous (proximal and distal) tumours has been challenged by recent data [15].
In addition to the finding that CpG island methylator phenotype (CIMP)-high tumours have particular phenotypical and prognostic features, they also evolve from a different precursor lesion than those tumours which are microsatellite stable (MSS) and show CIS. CIMP-high tumours form part of the serrated pathway and develop from precursor lesions, which have a different histological appearance to standard colonic adenomas [16, 17]. In addition to the known difference in prognosis for these tumours, there has also been the suggestion that they may also be associated with a lack of clinical benefit from standard 5-fluorouracil (5-FU)-based chemotherapy [18, 19].
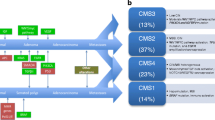
This molecular sub-typing of CRC has advanced to the stage where we can now begin to consider a molecular classification-based approach for CRC. Jass suggested one such classification and proposed five molecular sub-classifications (see Figure 1), based on levels of MSI and CIMP [17].
Molecular classification of CRC as described by Jass [17]. Tumours are divided primarily on the basis of CIMP status and microsatellite stability. Group 1 tumours show methylation of MLH1 and B-Raf mutations. They are characterised by CIMP+ and MSI-H, arise from serrated polyps and account for approximately 12% of CRC. Group 2 tumours are similar, but show only partial methylation of MSH1 associated with B-Raf mutation. They are CIMP+, MSS/microsatellite instable-low (MSI-L), arise from serrated polyps and account for approximately 8% of CRC. Group 3 tumours show not only mutations in APC, Kras and/or p53 but also methylation of MGMT. These tumours are CIMP-L and MSS/MSI-L, and show CIS. They can arise in either serrated or classical adenomas, and account for approximately 20% of CRC. Group 4 tumours are the classical type described in Vogelstein's original model [20], which show mutations in APC, Kras and/or p53. These tumours demonstrate CIS and are CIMP- and MSS. They arise in classical adenomas and make up approximately 57% of CRC. Group 5 tumours are those tumours arising in the familial cancer syndrome, HNPCC. They show mutations in one of the DNA MMR genes, are CIMP-, but MSI-H and account for approximately 3% of CRC (adapted from Ibrahim and Arends, [146]).
Introduction to current therapies
Four key approaches are currently used for the treatment of CRC: surgery, chemotherapy, radiotherapy and targeted therapies. The mainstay of CRC treatment is surgery. In early stage disease (stage 0 or I), surgical excision can be used without need for further treatment options, as the recurrence rate for node-negative T1 colorectal cancer is very low [21]. Many studies have now shown that adjuvant therapy has a survival benefit for patients with stage III disease, and therefore, this is the standard of care. The situation is not yet clear for patients with stage II CRC, however, in which there is somewhat conflicting evidence regarding the benefit of adjuvant therapy. It is agreed that ‘high-risk’ stage II patients should be offered adjuvant therapy, as they are the most likely to derive a benefit, although there is currently some debate regarding the exact definition of ‘high-risk’ stage II CRC. Patients with stage IV disease require chemotherapy or targeted therapies combined with surgery, where appropriate. Whilst surgery, chemotherapy and radiation therapy are key contributors to CRC treatment, the remainder of this article will focus on targeted therapies.
Targeted therapies in the treatment of CRC
As a consequence of improved understanding of the molecular pathology of cancer, a number of targeted agents have been developed which have demonstrated improved outcome in metastatic CRC (mCRC) patients, with combination chemotherapy. Among the first of these drugs to be developed and approved for use by the Food and Drug Administration (FDA) in mCRC were the following:
-
1.
Bevacizumab (Avastin™, Genentech/Roche, CA, USA), a monoclonal antibody targeted to vascular endothelial growth factor (VEGF) [22]; and
-
2.
The anti-EGFR monoclonal antibodies cetuximab (Erbitux™, Imclone Systems, NJ, USA [23]) and panitumumab (Vectibix™, Amgen, CA, USA [24]).
Anti-angiogenic drugs (bevacizumab)
Folkman et al. [25] first suggested the concept of targeting angiogenesis in cancer over 40 years ago. In 2004, the FDA approved bevacizumab as a first-line treatment for patients with mCRC. This followed a phase III study in 2004 on mCRC patients by Hurwitz et al. [22], which demonstrated that the inclusion of this drug to 5-FU combination therapy resulted in a ‘statistically significant and clinically meaningful improvement in survival’ among mCRC patients. Bevacizumab is a recombinant humanised monoclonal antibody that specifically targets VEGF-A, which is synthesised during tumour growth. Bevacizumab is thus defined as an anti-angiogenic drug due to its ability to prevent VEGF from interacting with appropriate receptors in vascular endothelial cells. As a result, cell signalling pathways that enhance angiogenesis, and thus the blood supply for tumours, are diminished. Bevacizumab is commonly used in combination with standard chemotherapeutic agents (e.g. 5-FU) as a first-line treatment for patients with mCRC and improves the overall survival (OS) of these patients by approximately 5 months [26, 27]. In theory, by specific targeting of endothelial cells, anti-angiogenic agents (such as bevacizumab) may avoid potential tumour cell resistance, thus indicating their usefulness in treating metastatic disease. Additionally, in contrast to EGFR-based therapies, bevacizumab is usually well tolerated by patients, and skin-based side effects are relatively rare [28]. This will be discussed in more detail later.
Limitations and side effects of bevacizumab
By targeting angiogenesis, a generalised biological phenomenon in the human body, bevacizumab-based therapies may result in toxicities affecting multiple organs. For example, in the Hurwitz et al. trial, a significantly higher rate of adverse effects was noted, including hypertension, anorexia and proteinuria [22]. A recent retrospective analysis of 228 Japanese patients with mCRC [29] showed that 96 (42%) did not receive bevacizumab as part of their treatment regime. In the majority of cases (76%), this was due to bevacizumab-specific contraindications, whilst an Italian study [30] demonstrated that 10% of mCRC patients had serious treatment-related toxicities. Of some concern is the correlation between gastrointestinal perforation, which occurs in 1%–2% of bevacizumab-treated patients, and mortality rates. This was reviewed recently in a community-based cohort of 1,953 CRC patients [31], and the authors found that most toxic events were successfully addressed by surgery. Other researchers have noted the limited survival benefits observed in some bevacizumab studies, whereby addition of bevacizumab to oxaliplatin-based chemotherapy significantly improved progression-free survival (PFS); however, OS differences did not reach statistical significance nor was response rate improved [32], whilst some preclinical and clinical studies have reported that VEGF inhibition actually increased tumour invasiveness and metastasis, suggesting that bevacizumab may indirectly stimulate new blood vessel growth (and tumour dissemination) through a VEGF-independent mechanism [33, 34].
Although considered rare, some cutaneous side effects of bevacizumab therapy have been observed, including exfoliative dermatitis, peripheral sensory neuropathy, skin discolouration and dryness [35]. However, it should also be noted that two studies by researchers at Yale University have suggested a potential association between skin rash occurrence in bevacizumab-treated mCRC patients and a positive drug response [36, 37], whilst Manzoni et al. [38] have suggested a role for the increased presence of circulating endothelial cells (CEC) as future predictive biomarkers for bevacizumab-treated mCRC patients.
Biomarkers and response to bevacizumab—potential predictive markers?
There is evidence that indicates that biomarkers may be of value in determining response to bevacizumab, as mentioned above. Such markers include vascular imaging, hypertension and polymorphisms affecting components of the VEGF pathway. In addition, profiles of circulating cytokines, growth factors and angiogenesis-related molecules may also prove valuable as prognostic and/or predictive markers.
Use of imaging tools
Imaging approaches, such as dynamic contrast enhanced (DCE)-magnetic resonance imaging (MRI) and 18F-fluorothymidine (FLT)-positron emission tomography (PET), may be used to assess the anti-angiogenic activity of bevacizumab. Small studies undertaken in advanced biliary tract cancer [39, 40] and breast cancer [41] have indicated that such approaches are promising as potential predictors of response. In the malignant glioma study, FLT-PET at 1–2 and 6 weeks was found to be a positive predictor of a survival benefit, whilst in biliary tract cancers, changes in FLT-PET (after 2 cycles of bevacizumab) were found to be a significant predictor of PFS and OS. In breast cancer patients receiving neoadjuvant chemotherapy plus bevacizumab, DCE-MRI indicated that greater decreases in angiogenic volume were associated with patients who derived a clinical benefit [42]. Whilst data in the CRC setting is more limited, there have been some indications that DCE-MRI may predict tumour shrinkage in response to combined bevacizumab and cytotoxic chemotherapy in CRC liver metastases [43]. However, further assessments using larger cohorts are necessary to determine if this approach has future value as a predictive biomarker of response to treatment.
Hypertension as a potential biomarker
Recent studies have indicated that the development of bevacizumab-induced arterial hypertension may serve as a potential predictive biomarker. Clinically, patients treated with bevacizumab have shown a rapid rise in blood pressure, and 5–18% have experienced grade 3 or 4 hypertension [44]. Hypertension, therefore, may be a useful biomarker of VEGF activity and predict the anti-angiogenic activity of bevacizumab. A number of small, single-arm studies have evaluated this in pancreatic cancer [45], renal carcinoma [46] and in CRC [47]. These studies have indicated that bevacizumab-induced hypertension (or the necessity for increased antihypertensive medication during bevacizumab treatment) was associated with extended PFS or OS. Further analysis carried out retrospectively on phase III trials in metastatic breast cancer and non-squamous, non-small cell lung cancer provides support for the biomarker as a predictive marker of clinical benefit [48, 49]. In CRC, however, the situation is less clear. Hurwitz et al. [50] undertook a retrospective analysis of two CRC studies and found that in only one of these studies did hypertensive changes (during bevacizumab therapy) predict a clinical benefit, as measured by PFS and OS, whilst Dewdney et al. [51] undertook a phase II study of bevacizumab-induced hypertension of 45 patients with poor-risk colorectal liver-only metastases. Although 15% of patients developed ≥ grade 1 hypertension (whilst receiving neoadjuvantx chemotherapy), and 4% developed grade 3 hypertension, no correlation was found between this condition and radiological response rate, PFS or OS.
This uncertainty could indicate that the predictive value of bevacizumab-induced hypertension might not extend to all cancers or treatment regimens, and thus, further work on CRC in large scale, prospective studies is required to elucidate the role of this as a potential biomarker.
Circulating biomarkers
Baseline measurements of circulating VEGF levels have been shown to be prognostic in a number of tumour types, including mCRC, lung cancer and renal-cell cancer. These studies also indicated that circulating VEGF levels were not predictive of treatment response to bevacizumab-based treatment regimes [52]. A recent study undertaken by Duda et al. [53] looked at the concentrations of VEGF, placental growth factor (PlGF), soluble VEGF receptor 1 (sVEGFR-1) and sVEGFR-2. These were measured in plasma and urine at baseline and during treatment in patients with locally advanced rectal cancer. In this study, they found that pre-treatment plasma sVEGFR-1, an endogenous blocker of VEGF and PlGF, and a factor linked with ‘vascular normalisation’ were associated with both primary tumour regression and the development of adverse events after neoadjuvant bevacizumab and chemoradiation [53].
A number of studies have been undertaken in an attempt to identify other circulating prognostic or predictive markers of bevacizumab therapy. A review of these studies can be found in Jubb and Harris [54] and by Wilson et al. [55]. In a study of 32 mCRC patients, Abajo et al. [56] utilised ELISAs and multiplex bead assays to determine if serum cytokines were predictive of bevacizumab efficacy. This group noted that high baseline serum levels of EGF and macrophage-derived chemokine and low levels of interleukin-6 (IL-6), IL-8 and IL-10 were correlated with likelihood of improved response. As mentioned previously, work by Manzoni et al. [38] on a group of 24 mCRC patients (undergoing bevacizumab treatment) indicated increased CECs and the apoptotic fraction of CECs as future (mutually independent) predictive biomarkers for this mCRC population, and appear to confirm other similar studies on CECs [57–61].
Polymorphisms in the VEGF pathway
Interest in the role of genetic variants in the VEGF pathway, as biomarkers for bevacizumab treatment outcome, has increased in light of the finding that heritability accounts for almost 80% of the variance seen in VEGF levels [62]. In metastatic breast cancer, Schneider et al. [48] identified two VEGF genotypes (VEGF-2578AA and VEGF-1154AA) which were significantly associated with improved OS in the bevacizumab plus paclitaxel treatment group. However, the same polymorphisms did not show any association with a PFS benefit. Both of these polymorphisms have also been associated with OS in a retrospective study of patients with mCRC receiving either FOLFIRI (leucovorin, fluorouracil, and irinotecan) plus bevacizumab, or XELIRI (capecitabine and irinotecan) plus bevacizumab [63]. A study undertaken by Lambrechts et al. [64] identified a locus in VEGFR1, which correlates with increased VEGFR1 expression and poor outcome of bevacizumab-treated patients with metastatic pancreatic adenocarcinoma and metastatic renal-cell carcinoma, although no work has as yet been undertaken in the CRC setting.
Despite the observations regarding the above polymorphisms, no data exist to indicate the biological significance of these variants and their role in response to treatment. As a result, additional work would provide a better understanding of their relevance.
Anti-EGFR therapy
EGFR is a 170 kD glycoprotein located in chromosome 7 [65] and is a member of the transmembrane tyrosine kinase receptor family, ErbB. EGFR is known to be over-expressed in tumours of epithelial origin, including CRC. EGFR activation is triggered by binding of peptide growth factors (of the EGF family) to the extracellular domain of the receptor and subsequently initiates both the RAS/RAF/MAPK and the phosphatidylinositol 3-kinase (PI3K signalling pathways. As a result, EGFR is directly involved in cell proliferation and survival, and therefore contributes to metastatic progression. Two classes of anti-EGFR therapies exist. The first group is composed of monoclonal antibodies to EGFR, whilst the second class includes tyrosine kinase inhibitors (TKIs), such as erlotinib and gefitinib.
Tyrosine kinase inhibitors
TKIs serve to block the cascade of reactions crucial to tumour development and survival; therefore, their anticancer properties have become an important focus for drug development by the pharmaceutical industry. To date, TKIs have obtained FDA approval for use in a range of cancers, including chronic myeloid leukaemia, acute lymphocytic leukaemia, non-small cell lung cancer and pancreatic cancer. Moreover, the use of the TKIs sunitinib, sorafenib and pazopanib into clinical care has doubled the PFS of patients with renal-cell cancer [66, 67]. As a result, sorafenib (originally developed as a BRAF inhibitor) is currently being tested in phase II trials for mCRC. Whilst current phase III trials of novel targeted agents for mCRC (specifically the small molecule TKIs, erlotinib and gefitinib) have been approved for targeting several cancers, neither has yet been approved for treating CRC. Additionally, the TKI imatinib has been used to treat advanced gastrointestinal stromal tumours; however, these tumours represent a very small proportion (<3%) of GI malignancies, are thus classed in a different category to CRC and are termed as connective tissue tumours.
Recent work has indicated the ability of TKIs to strongly suppress PI3K signalling (and possibly other pro-survival pathways) in CRC, suggesting that these treatments may have great therapeutic value [68]. Despite this, however, there has been no FDA approval granted to TKIs as new targeted agents for the treatment of mCRC since 2004. As a consequence, we will confine our discussions of anti-EGFR therapies to monoclonal antibodies.
Monoclonal antibodies to EGFR
Two EGFR antagonists, cetuximab (Erbitux™, Imclone Systems) and panitumumab (Vectibix™, Amgen) were FDA-approved for the treatment of mCRC in 2004 and 2006, respectively. Cetuximab is a chimeric immunoglobulin IgG1 monoclonal antibody that targets the ligand-binding domain of EGFR, whilst panitumumab is a human IgG2 monoclonal antibody. Both antibodies prevent EGFR autophosphorylation by binding to the extracellular domain and thus inhibiting activation of the downstream cell signalling pathways, MAPK and PI3K. Each antibody has been approved for the treatment of advanced CRC based on various parameters, including QoL, PFS and OS when used individually or in combination with standard chemotherapeutics.
Cetuximab
An early phase II trial that assessed the efficacy of cetuximab [69] by treatment-refractory CRC patients demonstrated the drug to have ‘modest activity’ that was well tolerated by patients. This study also noted that presence and severity of drug-induced acneiform skin rash correlated positively with survival [69]. Whilst in a seminal study of 329 mCRC patients, Cunningham et al. [70] showed that cetuximab had ‘clinically significant activity’ in patients with irinotecan-refractory CRC when given alone or in combination with this drug. Combination therapy patients demonstrated a higher response rate, longer median progression times and longer median survival times. Similarly, in a phase II trial of cetuximab by Vincenzi et al. [71], this anti-EGFR therapy was found to be clinically active in mCRC patients who were refractory to other regimens (irinotecan and oxaliplatin). In 2006, a multicentre study by Lenz et al. [72] demonstrated that cetuximab was well tolerated by mCRC patients who were previously unresponsive to irinotecan, oxaliplatin and fluoropyrimidines. As in previous studies, skin rash severity was correlated with drug response. Jonker et al. [73] undertook a study of cetuximab in treatment-resistant mCRC patients and noted that this drug therapy improved OS, PFS and preserved QoL measures, whilst a rash of grade 2 or above was robustly associated with improved survival.
Panitumumab
Following on from the successful use of cetuximab, a second anti-EGFR antibody, panitumumab, was introduced. Similar to cetuximab, panitumumab demonstrated significant anti-tumour activity in treatment-refractory mCRC patients, as well as a good safety profile. Studies of panitumumab by Gibson et al. [74] in a randomised phase III trial also demonstrated a reduction in tumour progression (46%) of patients with mCRC. Improved OS was positively correlated with severity of an acneiform skin rash, which was observed in 90% of subjects undergoing this treatment regime. Further, in a randomised phase III trial comparing panitumumab with other treatment regimens, including bevacizumab, Wainberg and Hecht [75] concluded that this therapy had ‘significant activity’ when used alone and improved PFS when compared with best supportive care (BSC). This finding was also observed in a study on treatment-refractory mCRC patients by Van Cutsem et al. [76]. Moreover, response rates of 10% were observed in panitumumab-treated subjects versus none for BSC. Yet, it should be noted that in this study, there was no change in OS. In a randomised phase III trial, Siena et al. [77] showed improved PFS in panitumumab-treated mCRC patients who were previously unresponsive to other regimens. Therefore, as a consequence of its success in addressing mCRC, panitumumab was approved by the FDA for use in the treatment of CRC and lung cancer.
Limitations and side effects of EGFR therapies
Skin toxicities are a common adverse event associated with EGFR inhibitors, with up to 80% of patients presenting with a skin rash (acneiform) on the scalp, face, neck and upper trunk [78]. This rash tends to occur within 3 weeks after therapy initiation, and it has been demonstrated that this response is reversible upon therapy cessation. In general, however, most studies suggest that EGFR therapies are well tolerated by patients. Both cetuximab and panitumumab have shown acceptable safety profiles in patients with mCRC. For example, Mitchell et al. [79] recently demonstrated that panitumumab (combined with irinotecan) had an acceptable toxicity profile in mCRC patients. Although in some cases, hypersensitivity reactions have been observed with cetuximab treatment [80, 81]. By contrast, hypersensitivity reactions are rare with panitumumab which has been indicated as an alternative treatment for patients at risk from cetuximab-induced anaphylaxis.
Biomarkers and response to anti-EGFR therapy—potential predictive markers?
Skin toxicity as a biomarker of response to anti-EGFR therapy
Research has suggested that adverse skin events following anti-EGFR therapies may be indicative of patient response. For example, cetuximab-induced skin rash in mCRC patients has been strongly correlated with improved PFS, OS and response rates in a number of studies [69, 82, 83]. Additionally, panitumumab has demonstrated a strong association between skin toxicities and improved outcome [84, 85]. Further pre-emptive [86] and prospective toxicity studies will enable researchers to clarify the mechanisms underlying the skin rash response in these therapies.
Major progress has been made using anti-EGFR therapies, including the understanding of biomarkers and their contribution to drug response. Despite the success of anti-EGFR therapy, however, not all patients obtain the same benefits. This has led to the search for biomarkers which might indicate the patient population that would benefit most from anti-EGFR therapy. As such therapies are expensive and potentially toxic, the search for such biomarkers is driven, not only from the clinical and scientific community, but also from the payor community. Therefore, specific targeting of patients who would benefit from this treatment is being actively pursued.
EGFR polymorphisms/mutations and overexpression
To date, a small number of pharmacogenetic studies have identified EGFR polymorphisms as potential biomarkers in predicting response to anti-EGFR therapies in mCRC and non-small cell lung cancer [87–89]. By contrast to lung adenocarcinoma, however, EGFR mutations in CRC are somewhat rare [90–92], thus limiting their utility as biomarkers at present.
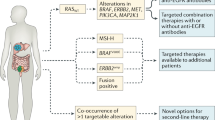
Immunohistochemical (IHC) studies of CRC tumours indicated that patients with EGFR-positive tumours benefit from anti-EGFR therapy [93]. Since 2004, the combination of a diagnostic test for EGFR (EGFR PharmDx™ Kit, Dako, Cambridgeshire, UK), and the subsequent application of cetuximab or panitumumab in EGFR-positive colon carcinomas has been approved by FDA, reviewed in [94]. However, subsequent studies have shown that EGFR-negative tumours may also benefit from cetuximab therapy [69, 95, 96]. These studies appear to indicate that analysis of EGFR expression via IHC may not be as reliable in predicting the efficacy of EGFR therapy. As a consequence of these disappointing results, researchers have instead undertaken a search for alternative biomarkers. These investigations have lead to a focus on downstream effectors of EGFR signalling, including RAS family members, BRAF and the PI3K pathway, amongst others. Each of these potential biomarkers will be discussed individually.
-
1.
KRAS
KRAS is a member of the RAS proto-oncogene family which is transiently activated by the action of ligand binding of EGFR. The RAS family also includes NRAS and HRAS. KRAS mutations have been reported in approximately 40% of human CRC [97–100]. The majority (85%) of these mutations occur in codons 12 and 13, with a smaller number occurring in codons 61, 117 and 146 [101–103]. Oncogenic mutation in KRAS renders the protein constitutively active, thus maintaining it in the GTP-bound conformation.
Due to the common occurrence of KRAS mutations in CRC, a number of studies have examined their potential clinical relevance. Some studies have shown that the presence of a KRAS mutation may have prognostic significance, as reviewed in [92]. This effect appears to be borderline and confined to certain mutations (particularly the G12V mutation). Moreover, the effect may be confined to certain stages of CRC.
The importance of KRAS mutation was truly realised with the advent of anti-EGFR monoclonal antibodies as a treatment for mCRC. These were introduced into clinical use around 2004 and showed reasonable response rates of 10%–15% in mCRC. However, their role was revolutionised by the discovery that patients whose tumours harbour mutant forms of KRAS do not respond to anti-EGFR therapy [104]. Although the results of this study were statistically significant, patient numbers were small and there was a need for greater sample sizes. Subsequent studies with larger numbers confirmed these findings [85, 105]. Meta-analyses of the available randomised controlled trials have gone on to further support these results, demonstrating that a benefit from anti-EGFR therapy is only seen in patients whose tumour is wild-type for KRAS[106–108]. In these studies, no difference was found in the predictive effects of mutations in different codons.
Based on these and other studies, the American Society of Clinical Oncology recommended that patients who are candidates for anti-EGFR therapy should have their tumour tested for KRAS mutation. If a codon 12 or 13 mutation is detected, these patients should not be treated with anti-EGFR therapy [109]. Similarly, the Canadian Expert Group has stated that KRAS status should be determined whenever anti-EGFR therapy is being considered in the setting of mCRC [110, 111]. In 2009, the FDA updated the product labels for cetuximab and panitumumab, indicating that patients with CRC tumours harbouring KRAS mutations were unlikely to derive benefit from these therapies. However, these guidelines may have to be re-visited, as it has been shown (in one study) that patients with a G13D mutation showed improved survival compared to patients with other mutations, indicating that this patient group may respond to therapy [112]. This is, therefore, an area that will require further study.
-
2.
KRAS mutation heterogeneity
Although there is now widespread agreement that the presence of a KRAS mutation indicates that a patient is unlikely to respond to anti-EGFR therapy, there is some concern around the area of tumour heterogeneity. Some studies have shown a strong correlation in mutation status between primary tumours and distant metastases [113, 114]. Yet, other studies have suggested that there is significant heterogeneity in the expression of KRAS mutations within a tumour [115–118] and also between primary tumour samples and lymph node metastases [113, 117]. This could theoretically lead to false negative results, i.e. there is a KRAS mutation present within a tumour, but the portion sampled for testing does not contain the mutation (or there are insufficient numbers of tumour cells with the mutation) to enable detection. Whilst this issue has been raised in the literature, to date, no reliable data have been provided to suggest that appropriate sampling protocols are in place. A robust sampling method would address this concern [118].
-
3.
Approach used for KRAS mutation testing
The analysis of tumour samples for KRAS mutations is undertaken using samples of fresh-frozen or paraffin-embedded tumour material. A pathology review of the material is essential to establish that the sample contains sufficient tumour cells for analysis. If necessary, enrichment for tumour material may be carried out using tumour microdissection. DNA is then extracted from the sample and used for molecular testing using sequencing or pyrosequencing-based approaches, or an allele-specific PCR-based approach. Currently, there are no FDA-approved protocols for KRAS testing. However, a number of laboratory-based assays can be utilised, provided they are run under CAP certification or other CMS-approved certification (as required by the CLIA regulations of 1988). In countries where CE marking is accepted, there are a number of approved KRAS mutation tests available for use in CRC. A summary of these tests is listed in Table 1 below.
-
4.
The cost/benefit ratio of KRAS testing
The use of mutation testing has obvious benefits to patients by preventing exposure of those patients unlikely to respond, to potential toxic effects of a given drug regime. Moreover, there are significant economic benefits. Studies have demonstrated that the investment in testing for KRAS and BRAF (discussed later) results in significant cost savings [119, 120]. For example, as early as 2009, researchers were estimating that routine testing for KRAS mutations in CRC patients would save approximately $740 million a year, whilst more recently, Vijayaraghavan et al. [121] have estimated that KRAS mutation testing saves $7,500–$12,400 per patient in the USA and €3,900–€9,600 per patient in Germany.
Other potential biomarkers of mCRC
The finding of a link between KRAS mutation status and anti-EGFR therapy has led to the investigation of other downstream effectors as potential biomarkers. The most investigated candidates have been NRAS, BRAF, PI3K and phosphatase and tensin homologue (PTEN). Each of these will be dealt with in more detail in the next section.
-
1.
NRAS
Mutations in NRAS are less common than KRAS and are present in approximately 2.6% of CRC cases [112]. As a result of this infrequency, only a limited number of studies investigating the predictive potential of NRAS have been undertaken. De Rooke et al. [112] showed a significant difference in response rate between patients in whom tumours showed an NRAS mutation (1/13, 7.7%) versus patients whose tumour was wild-type NRAS (110/289, 38.1%). Although, it should be noted that no significant difference in disease control rate, PFS or OS was observed.
-
2.
BRAF
V-raf murine sarcoma viral oncogene homologue B1 (BRAF) is a serine-threonine kinase, which is downstream of RAS. BRAF is an important oncogene and is found to be mutated in a number of malignancies, including sporadic CRC, where the rate of mutation is of the order of 15% [108]. BRAF mutation has been associated with the serrated pathway of tumour development, and these tumours tend to be in groups 1 and 2 of the Jass classification [17]. The vast majority of BRAF mutations in CRC are the V600E mutation, which leads to constitutive activation of the kinase [122].
A number of the earlier studies examining biomarkers associated with a response to anti-EGFR therapy did not contain any CRC patients with a BRAF mutation. This has generally been a problem, where low patient numbers have led to poor statistical power. However, more recent research has overcome this, and studies have generally shown that the presence of a BRAF mutation correlates negatively with response to anti-EGFR therapy [98, 112, 123, 124]. In a cohort of 173 CRC patients, Laurent-Puig et al. [123] noted that five patients with a BRAF mutation showed a significant decrease in PFS and OS when compared to BRAF wild-type patients treated with cetuximab. Although the numbers were small, this study did demonstrate statistical significance. De Roock et al. [112] have reported the largest cohort of BRAF mutant tumours to date with 36/761 (4.7%) of tumours tested containing a BRAF mutation. This group also demonstrated a significant difference in response rate between BRAF mutant and wild-type tumours.
Lin et al. [106] recently performed a systematic review of the predictive value of BRAF in mCRC. They concluded that, although the quality of the studies is impaired by their retrospective nature, there is definite evidence supporting the view that mutation of BRAF predicts a lack of response to anti-EGFR therapy. A subsequent meta-analysis by Mao et al. [125] has supported the results of these individual studies. This study observed that in KRAS wild-type patients, the objective response to anti-EGFR therapy was 0% in BRAF mutant tumours versus 36.3% in BRAF wild-type tumours. Further, Prahallad et al. [126] have recently suggested that BRAF (V600E) mutant colon cancers may benefit from a combined therapy of BRAF and EGFR inhibitors.
The data has now developed to the point where testing for BRAF is becoming part of the clinical routine for the management of mCRC patients in some centres. BRAF mutation testing is being incorporated into commercially available test kits for this purpose. It has also been shown that KRAS and BRAF analyses together have a favourable cost-benefit profile [119].
-
3.
PIK3CA
Phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha, PIK3CA, is a phospholipid kinase, which is downstream of EGFR but signals via AKT rather than via the canonical MAP kinase pathway. As with KRAS and BRAF, PIK3CA undergoes activating mutations. The most common of these occurs in either exon 9 (approximately 60%, G1624A; E542K) or exon 20 (approximately 25%, A3140G; H1047R). These mutations result in different effects. Both are activating mutations, but exon 9 mutations require an interaction with activated RAS for subsequent downstream signalling. In the case of the exon 20 mutation, downstream signalling is independent of RAS [127].
There have been some suggestions that, in an in vitro setting at least, this may be the more important pathway in terms of the oncogenic effect of EGFR signalling [128]. Therefore, there has been significant interest in whether components of this pathway will impact on response to anti-EGFR therapy. As with BRAF studies, small cohort sizes have hampered attempts to definitively determine the predictive value of PIK3CA mutations.
There has been some conflicting clinical data regarding the importance of PI3K mutations in predicting response to anti-EGFR therapy. In their initial study, Lièvre et al. [104] did not show any correlation between PIK3CA mutation and response to cetuximab, but their study only included three patients with a PIK3CA mutation. A larger study (containing 15 tumours with a PIK3CA mutation) by Sartore-Bianchi et al. [129] showed a significant association between mutation of the gene and resistance to cetuximab or panitumumab. Similar results were obtained by Souglakos et al. [98]. However, these results were not replicated by others [130]. These differing results may have been reconciled by the work of De Roock et al. [112]. They [112] showed that exon 9 mutations had no effect on response to treatment, whilst exon 20 mutations were significantly correlated with objective response, PFS and OS. In their study, no patient with an exon 20 mutation responded to cetuximab-based therapy. A recent meta-analysis has supported these results, showing an objective response rate of 0% in patients whose tumours harboured a PIK3CA exon 20 mutation [125]. By contrast, the objective response rate was found to be 37% in patients with tumours that were wild-type for PIK3CA exon 20.
-
4.
PTEN
PTEN is a key inhibitor of the PIK signalling pathway, and its expression has been shown to be lost in a number of different human tumours, including CRC. The mechanism of decreased expression can be driven by mutation or epigenetic phenomena, with up 13% of CRC in the COSMIC database showing a PTEN mutation. It has been suggested that this latter mechanism is particularly prevalent in tumours with high levels of MSI [131].
A number of CRC studies have examined PTEN expression and response to anti-EGFR therapy. These studies have primarily used IHC to assess expression at the protein level, and some have shown a correlation between PTEN expression and clinical response [123, 132–134]. However, other studies have not shown such a relationship [135–137]. For example, Loupakis et al. [138] showed that PTEN expression in metastatic, but not in primary, tumours correlated with response to cetuximab. Moreover, Negri et al. [139] used indirect immunofluorescence to determine PTEN loss and showed that none of the patients with PTEN loss responded to cetuximab therapy. This was compared to a 30% disease progression rate in patients whose tumour retained PTEN expression [124]. These differing results may be due to the method of determining PTEN loss, i.e. IHC. Moreover, the studies examining PTEN have used differing protocols, antibodies and scoring systems, and this is reflected in the differing number of tumours found to be PTEN-deficient. This conflicting evidence has led the authors of at least one systematic review to conclude that, at present, PTEN expression levels cannot robustly predict response to anti-EGFR therapy [106]. Therefore, until there is further study in this area and until a standardised approach to assessment can be implemented, it appears that PTEN cannot be used in routine clinical practice.
-
5.
pAKT
Closely associated with PIK3CA and PTEN testing, is activated AKT (pAKT). The presence of pAKT indicates activation of the pathway and therefore may indicate PIK3CA mutations, PTEN loss or activation of the pathway via a different mechanism. A number of studies have assessed the effect of pAKT expression in CRC and attempted to correlate it with response to anti-EGFR therapy. To date, however, no significant correlation has been found [138, 140, 141].
Drug/diagnostic co-development—future approaches for personalised medicine
With the advance of our knowledge around patient segmentation, it will be necessary to consider the development of a companion diagnostic to aid treatment decision making during the drug development process. Ideally, the development of the diagnostic and the drug should happen in parallel, and the regulatory approval of both should be achieved simultaneously. Recent US FDA draft guidance on the development of companion diagnostics has been published [142]. In this draft guidance, the FDA sets forth its expectations for the development of companion diagnostic tools that will be used to determine treatment choices. This document also outlines the requirement for drug development and diagnostic tools to be carried out simultaneously. However, as recent events have indicated, this situation is not always achievable. In many circumstances, the understanding of the biology of the action of the drug in the intended disease population is evolving. Therefore, the knowledge of which biomarkers are important for the selection of appropriate patients for treatment may not be clear early enough in the programme to make the development of a diagnostic (in time for launch) a realistic goal. This is illustrated with the understanding around the role of KRAS mutations in resistance to anti-EGFR monoclonal antibody treatment. It is evident from recent clinical data from the phases II and III trials for panitumumab and cetuximab, where patients whose tumours harbour mutations in KRAS are less likely to respond to treatment. Retrospective analyses across seven randomised clinical trials suggest that anti-EGFR-directed monoclonal antibodies are not effective for the treatment of patients with mCRC containing KRAS mutations. This finding has led to the inclusion of information on the FDA-approved drug label for panitumumab and cetuximab (Indications and Usage section) to the effect that treatment of patients whose tumours harbour KRAS mutations is not recommended. Currently, however, there is no FDA-approved companion KRAS mutation test for either drug. The key rationale for KRAS mutation testing in this patient population is to avoid the exposure of patients to unnecessary drug toxicities in situations where there is unlikely to be any clinical benefit. In addition, this avoids unnecessary financial burdens on the healthcare system. However, it does remain that CRC patients whose tumours harbour KRAS mutations have a high unmet medical need. Understanding the role of additional biomarkers in CRC and their potential use as predictive markers of drug response will show possible utility for the future and will allow us to see an increase in the development of companion diagnostic tools to support this.
Recent National Institute for Health and Clinical Excellence ruling on use of bevacizumab
The UK National Institute for Health and Clinical Excellence has recently published guidelines on the use of VEGF- and EGFR-targeted therapies in treating mCRC patients who have failed to respond adequately to first-line treatments [143]. The guidelines have acknowledged the fact that cetuximab showed benefits in terms of PFS and OS when used in KRAS wild-type mCRC patients compared with BSC alone. They also noted that panitumumab, given as a monotherapy, had shown a benefit in terms of PFS. However, following cost-benefit analysis, the committee concluded that cetuximab and panitumumab (in addition to bevacizumab) were ‘not a cost-effective use of National Health Service (NHS) resources’ [143]. This analysis highlights the need to continue to refine our strategies in terms of the use of biomarkers and to identify those patients who are likely to derive the most benefit from these treatments. This will not only promote genuine personalised medicine, but will also enable us to use these therapies in a manner that will not put undue strain on finite healthcare resources.
Conclusions
In recent years, significant steps have been made in the diagnosis and successful treatment of CRC. Researchers and clinicians, however, are still faced with challenges, not least the detection and management of tumours with varied gene mutation status. Further clarification of the molecular pathology of CRC may improve treatment options as well as improve QoL and ultimately, the long-term survival of patients with this condition. As new technologies emerge allowing the identification of CRC gene mutation status, this information will inform clinicians as to the most appropriate and efficacious form of treatment. This individualised approach to managing mCRC fits in well with the PPPM model put forward by EPMA [144, 145]. However, in order to gain most benefit from these novel technologies, targeted therapy must be informed by well-designed studies, large cohort sizes and internationally standardised detection protocols. Therefore, it is imperative that scientists and clinicians collaborate closely in developing new ways to deal with the vast quantities of data generated by mutational analyses in order to make sense of the underlying mechanisms of CRC. As human populations are living longer, healthcare systems around the world will be challenged to find new ways of dealing with CRC to improve global health.
Expert recommendations
The following are expert recommendations:
-
Routine KRAS and BRAF mutation testing of patients with mCRC who are being considered for anti-EGFR therapy to ensure they receive the most appropriate therapy whilst being spared exposure to non-beneficial therapeutic effects.
-
PIK3CA and PTEN have also shown promise as informative biomarkers for EGFR therapy, but further studies are necessary before they can be used clinically on a routine basis.
-
Panel-based biomarker testing platforms should be developed as a way to ensure that maximal value can be derived from limited clinical material.
-
VEGF inhibitors have shown promise in the treatment of mCRC, but more research is required into identifying predictive markers for patient response. This will allow a more personalised approach to anti-VEGF treatments in the future.
-
Access to high-quality tumour material for biomarker testing is of paramount importance; therefore, tissue collection and storage procedures must be standardised internationally.
-
Development of internationally accessible bio-banks for CRC tissues is necessary to progress high-quality biomarker research.
-
Good practice is essential in maintaining detailed patient records for developing PPPM, based on multifactorial data, e.g. sex, age, weight, ethnicity, lifestyle, and prior therapy regimens.
-
High stringency of genomic and proteomic assays is required across research labs to ensure reproducibility of data.
-
Early biomarker discovery, coupled with approaches to ensure appropriate co-development of formative companion diagnostics, should be a standard practice in all drug development programmes.
-
Improved clinical trial design with more stringent stratification of patient cohorts.
Authors’ information
SH (PhD FEA) is a lecturer in Anatomy and Physiology in the Department of Life Sciences at GCU, with a special interest in predictive and prognostic biomarkers of inflammation and disease. Her previous research background included publications on the metastasis suppressor gene, RKIP, in mCRC and breast cancer. MO is a Diagnostic Team Director in the Personalised Healthcare and Biomarker Function within AstraZeneca, where her role involves her in championing the use of biomarkers to support oncology drug development. BD (MB BAO BCh PhD FRCPath) is a lecturer in Histopathology at Trinity College Dublin, with a special interest in gastrointestinal and molecular pathology.
Abbreviations
- 5-FU:
-
5-fluorouracil
- BSC:
-
Best supportive care
- CIMP:
-
CpG island methylator phenotype
- CIS:
-
Chromosomal instability
- CRC:
-
Colorectal cancer
- DCE-MRI:
-
Dynamic contrast enhanced-magnetic resonance imaging
- EGFR:
-
Epidermal growth factor receptor
- FDA:
-
Food and Drugs Administration
- FLT-PET:
-
18F-fluorothymidine-positron emission tomography
- HNPCC:
-
Hereditary non-polyposis colorectal cancer
- IHC:
-
Immunohistochemistry
- mCRC:
-
Metastatic colorectal cancer
- MMR:
-
Mismatch repair
- MSI:
-
Microsatellite instability
- MSI-H:
-
High level microsatellite instability
- MSI-L:
-
Low level microsatellite instability
- MSS:
-
microsatellite stable
- NHS:
-
National Health Service (UK)
- OS:
-
Overall survival
- PFS:
-
Progression-free survival
- PI3K :
-
Phosphatidylinositol 3-kinase
- PlGF:
-
Placental growth factor
- PPPM:
-
Predictive, preventive and personalised medicine
- PTEN:
-
Phosphatase and tensin homologue
- QoL:
-
Quality of life
- sVEGFR-1:
-
Soluble VEGF receptor 1
- TKI:
-
Tyrosine kinase inhibitors
- VEGF:
-
Vascular endothelial growth factor.
References
Ferlay J, Parkin DM, Steliarova-Foucher E: Estimates of cancer incidence and mortality in Europe in 2008. Eur J Cancer. 2010, 46 (4): 765-781. 10.1016/j.ejca.2009.12.014.
Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, Dezso Z, Ustyanksky V, Nikolskaya T, Nikolsky Y, Karchin R, Wilson PA, Kaminker JS, Zhang Z, Croshaw R, Willis J, Dawson D, Shipitsin M, Willson JK, Sukumar S, Polyak K, Park BH, Pethiyagoda CL, Pant PV: The genomic landscapes of human breast and colorectal cancers. Science. 2007, 318 (5853): 1108-1113. 10.1126/science.1145720.
Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH: Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008, 321 (5897): 1801-1806. 10.1126/science.1164368.
Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G: An integrated genomic analysis of human glioblastoma multiforme. Science. 2008, 321 (5897): 1807-1812. 10.1126/science.1164382.
Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, Ivanova E, Watson IR, Nickerson E, Ghosh P, Zhang H, Zeid R, Ren X, Cibulskis K, Sivachenko AY, Wagle N, Sucker A, Sougnez C, Onofrio R, Ambrogio L, Auclair D, Fennell T, Carter SL, Drier Y, Stojanov P, Singer MA, Voet D, Jing R, Saksena G, Barretina J: Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012, 485 (7399): 502-506.
Wagle N, Berger MF, Davis MJ, Blumenstiel B, Defelice M, Pochanard P, Ducar M, Van Hummelen P, Macconaill LE, Hahn WC, Meyerson M, Gabriel SB, Garraway LA: High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012, 2 (1): 82-93. 10.1158/2159-8290.CD-11-0184.
McBride DJ, Etemadmoghadam D, Cooke SL, Alsop K, George J, Butler A, Cho J, Galappaththige D, Greenman C, Howarth KD, Lau KW, Ng CK, Raine K, Teague J, Wedge DC, Caubit X, Stratton MR, Brenton JD, Campbell PJ, Futreal PA, Bowtell DD, Australian Ovarian Cancer Study Group: Tandem duplication of chromosomal segments is common in ovarian and breast cancer genomes. J Pathol. 2012, 227 (4): 446-455. 10.1002/path.4042.
Migliore L, Migheli F, Spisni R, Coppedè F: Genetics, cytogenetics, and epigenetics of colorectal cancer. J Biomed Biotechnol. 2011, 2011: 792362-
Markman B, Javier Ramos F, Capdevila J, Tabernero J: EGFR and KRAS in colorectal cancer. Adv Clin Chem. 2010, 51: 71-119.
He J, Efron JE: Screening for colorectal cancer. Adv Surg. 2011, 45: 31-44. 10.1016/j.yasu.2011.03.006.
SEER Cancer Statistics Review, 1975–2009 (Vintage 2009 Populations). Edited by: Howlander N, Noone AM, Krapcho M, Neyman N, Aminou R, Altekruse SF, Kosary CL, Ruhl J, Tatalovich Z, Cho H, Mariotto A, Eisner MP, Lewis DR, Chen HS, Feuer EJ, Cronin KA. 2012, Bethesda: National Cancer Institute
National Cancer Intelligence Network (NCIN): Data Briefing. Colorectal Cancer Survival by Stage. [http://www.ncin.org.uk/publications/data_briefings/colorectal_cancer_survival_by_stage.aspx]
Pino MS, Chung DC: Microsatellite instability in the management of colorectal cancer. Expert Rev Gastroenterol Hepatol. 2011, 5 (3): 385-399. 10.1586/egh.11.25.
Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, Dietrich D, Biesmans B, Bodoky G, Barone C, Aranda E, Nordlinger B, Cisar L, Labianca R, Cunningham D, Van Cutsem E, Bosman F: Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60–00 trial. J Clin Oncol. 2010, 28 (3): 466-474. 10.1200/JCO.2009.23.3452.
Yamauchi M, Morikawa T, Kuchiba A, Imamura Y, Qian ZR, Nishihara R, Liao X, Waldron L, Hoshida Y, Huttenhower C, Chan AT, Giovannucci E, Fuchs C, Ogino S: Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012, 61 (6): 847-854. 10.1136/gutjnl-2011-300865.
Zlobec I, Bihl M, Foerster A, Rufle A, Lugli A: Comprehensive analysis of CpG island methylator phenotype (CIMP)-high, -low, and -negative colorectal cancers based on protein marker expression and molecular features. J Pathol. 2011, 225 (3): 336-343. 10.1002/path.2879.
Jass JR: Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007, 50 (1): 113-130. 10.1111/j.1365-2559.2006.02549.x.
Jover R, Nguyen TP, Pérez-Carbonell L, Zapater P, Payá A, Alenda C, Rojas E, Cubiella J, Balaguer F, Morillas JD, Clofent J, Bujanda L, Reñé JM, Bessa X, Xicola RM, Nicolás-Pérez D, Castells A, Andreu M, Llor X, Boland CR, Goel A: 5-fluorouracil adjuvant chemotherapy does not increase survival in patients with CpG island methylator phenotype colorectal cancer. Gastroenterology. 2011, 140 (4): 1174-1181. 10.1053/j.gastro.2010.12.035.
Des Guetz G, Uzzan B, Nicolas P, Schischmanoff O, Perret GY, Morere JF: Microsatellite instability does not predict the efficacy of chemotherapy in metastatic colorectal cancer: a systematic review and meta-analysis. Anticancer Res. 2009, 29 (5): 1615-1620.
Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL: Genetic alterations during colorectal-tumor development. N Engl J Med. 1988, 319 (9): 525-532. 10.1056/NEJM198809013190901.
Kobayashi H, Mochizuki H, Morita T, Kotake K, Teramoto T, Kameoka S, Saito Y, Takahashi K, Hase K, Oya M, Maeda K, Hirai T, Kameyama M, Shirouzu K, Sugihara K: Characteristics of recurrence after curative resection for T1 colorectal cancer: Japanese multicenter study. J Gastroenterol. 2011, 46 (2): 203-211. 10.1007/s00535-010-0341-2.
Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, Berlin J, Baron A, Griffing S, Holmgren E, Ferrara N, Fyfe G, Rogers B, Ross R, Kabbinavar F: Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Eng J Med. 2004, 350: 2335-2342. 10.1056/NEJMoa032691.
Van Cutsem E, Köhne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D'Haens G, Pintér T, Lim R, Bodoky G, Roh JK, Folprecht G, Ruff P, Stroh C, Tejpar S, Schlichting M, Nippgen J, Rougier P: Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009, 360: 1408-1417. 10.1056/NEJMoa0805019.
Douillard JY, Siena S, Cassidy J, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, Rivera F, Kocákova I, Ruff P, Błasińska-Morawiec M, Šmakal M, Canon JL, Rother M, Oliner KS, Wolf M, Gansert J: Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol. 2010, 28: 4697-4705. 10.1200/JCO.2009.27.4860.
Folkman J, Merler E, Abernathy C, Williams G: Isolation of a tumor factor responsible for angiogenesis. J Exp Med. 1971, 133 (2): 275-288. 10.1084/jem.133.2.275.
Samelis GF, Ekmektzoglou KA, Tsiakou A, Konstadoulakis M: The continuation of bevacizumab following disease progression in patients with metastatic colorectal cancer offers a survival benefit. Hepatogastroenterology. 2011, 58 (112): 1968-1971.
Guan ZZ, Xu JM, Luo RC, Feng FY, Wang LW, Shen L, Yu SY, Ba Y, Liang J, Wang D, Qin SK, Wang JJ, He J, Qi C, Xu RH: Efficacy and safety of bevacizumab plus chemotherapy in Chinese patients with metastatic colorectal cancer: a randomized phase III ARTIST trial. Chin J Cancer. 2011, 30 (10): 682-689. 10.5732/cjc.011.10188.
Zoratto F, Rossi L, Zullo A, Papa A, Zaccarelli E, Tomao L, Giordani E, Colonna M, Baiano G, Tomao S: Critical appraisal of bevacizumab in the treatment of metastatic colorectal cancer. Onco Targets Ther. 2012, 5: 199-211.
Hori N, Iwasa S, Hashimoto H, Yanai T, Kato K, Hamaguchi T, Yamada Y, Murakoshi K, Yokote N, Yamamoto H, Shimada Y: Reasons for avoidance of bevacizumab with first-line FOLFOX for advanced colorectal cancer. Int J Clin Oncol. 10.1007/s10147-012-0398-4.
Bonifazi M, Rossi M, Moja L, Scigliano VD, Franchi M, La Vecchia C, Zocchetti C, Negri E: Bevacizumab in clinical practice: prescribing appropriateness relative to national indications and safety. Oncologist. 2012, 17 (1): 117-124. 10.1634/theoncologist.2011-0184.
Kabbinavar FF, Flynn PJ, Kozloff M, Ashby MA, Sing A, Barr CE, Grothey A: Gastrointestinal perforation associated with bevacizumab use in metastatic colorectal cancer: results from a large treatment observational cohort study. Eur J Cancer. 2012, 48 (8): 1126-1132. 10.1016/j.ejca.2012.02.052.
Saltz LB, Clarke S, Díaz-Rubio E, Scheithauer WA, Wong R, Koski S, Lichinitser M, Yang TS, Rivera F, Couture F, Sirzén F, Cassidy J: Bevacizumab in combination with oxaliplatin-based chemotherapy as first-line therapy in metastatic colorectal cancer: a randomized phase III study. J Clin Oncol. 2008, 26 (12): 2013-2019. 10.1200/JCO.2007.14.9930.
Bergers G, Hanahan D: Modes of resistance to anti-angiogenic therapy. Nat Rev Cancer. 2008, 8 (8): 592-603. 10.1038/nrc2442.
Leite De Oliveira R, Hamm A, Mazzone M: Growing tumor vessels: more than one way to skin a cat - implications for angiogenesis targeted cancer therapies. Mol Aspects Med. 2011, 32 (2): 71-87. 10.1016/j.mam.2011.04.001.
Wozel G, Sticherling M, Schön MP: Cutaneous side effects of inhibition of VEGF signal transduction. J Dtsch Dermatol Ges. 2010, 8 (4): 243-249. 10.1111/j.1610-0387.2009.07268_supp.x.
Gotlib V, Khaled S, Lapko I, Mar N, Saif MW: Skin rash secondary to bevacizumab in a patient with advanced colorectal cancer and relation to response. Anticancer Drugs. 2006, 17 (10): 1227-1229. 10.1097/01.cad.0000231481.07654.fc.
Saif MW, Longo WL, Israel G: Correlation between rash and a positive drug response associated with bevacizumab in a patient with advanced colorectal cancer. Clin Colorectal Cancer. 2008, 7 (2): 144-148. 10.3816/CCC.2008.n.020.
Manzoni M, Mariucci S, Delfanti S, Rovati B, Ronzoni M, Loupakis F, Brugnatelli S, Tinelli C, Villa E, Falcone A, Danova M: Circulating endothelial cells and their apoptotic fraction are mutually independent predictive biomarkers in bevacizumab-based treatment for advanced colorectal cancer. J Cancer Res Clin Oncol. 2012, 138 (7): 1187-1196. 10.1007/s00432-012-1190-6.
Zhu AX, Meyerhardt JA, Blaszkowsky LS, Kambadakone AR, Muzikansky A, Zheng H, Clark JW, Abrams TA, Chan JA, Enzinger PC, Bhargava P, Kwak EL, Allen JN, Jain SR, Stuart K, Horgan K, Sheehan S, Fuchs CS, Ryan DP, Sahani DV: Efficacy and safety of gemcitabine, oxaliplatin, and bevacizumab in advanced biliary-tract cancers and correlation of changes in 18-fluorodeoxyglucose PET with clinical outcome: a phase 2 study. Lancet Oncol. 2010, 11: 48-54. 10.1016/S1470-2045(09)70333-X.
Chen W, Delaloye S, Silverman DH, Geist C, Czernin J, Sayre J, Satyamurthy N, Pope W, Lai A, Phelps ME, Cloughesy T: Predicting treatment response of malignant gliomas to bevacizumab and irinotecan by imaging proliferation with [18F] fluorothymidine positron emission tomography: a pilot study. J Clin Oncol. 2007, 25: 4714-4721. 10.1200/JCO.2006.10.5825.
Mrozek E, Lustberg MB, Knopp MV, Spigos DG, Yang X, Houton LA, Ramaswamy B, Layman RM, Povoski SP, Agnese DM: Phase II trial of neoadjuvant chemotherapy (NCT) with weekly nanoparticle albumin-bound paclitaxel (Nab-P), carboplatin (CBP), and bevacizumab (BEV) in women with clinical stages II-III breast cancer (BC): pathologic response prediction by changes in angiogenic volume (AV) by dynamic contrast magnetic resonance imaging (DCE-MRI) [abstract]. J Clin Oncol. 2010, 28: 604-
Zweifel M, Padhani AR: Perfusion MRI in the early clinical development of antivascular drugs: decorations or decision making tools?. Eur J Nucl Med Mol Imaging. 2010, 37 (Suppl 1): S164-S182.
O'Connor JPB, Rose CJ, Jackson A, Watson Y, Cheung S, Maders F, Whitcher BJ, Roberts C, Buonaccorsi GA, Thompson X, Clamp AR, Jayson GC, Parker GJM: DCE-MRI biomarkers of tumour heterogeneity predict CRC liver metastasis shrinkage following bevacizumab and FOLFOX-6. Br J Cancer. 2011, 105: 139-145. 10.1038/bjc.2011.191.
Syrigos KN, Karapanagiotou E, Boura P, Manegold C, Harrington K: Bevacizumab-induced hypertension: pathogenesis and management. BioDrugs. 2011, 25 (3): 159-169. 10.2165/11590180-000000000-00000.
Friberg G, Kasza K, Vokes EE, Kindler HL: Early hypertension (HTN) as a potential pharmacodynamic (PD) marker for survival in pancreatic cancer (PC) patients (pts) treated with bevacizumab (B) and gemcitabine (G) [abstract]. J Clin Oncol. 2005, 23: 3020-
Bono P, Elfving H, Utriainen T, Osterlund P, Saarto T, Alanko T, Joensuu H: Hypertension and clinical benefit of bevacizumab in the treatment of advanced renal cell carcinoma. Ann Oncol. 2009, 20: 393-394.
Scartozzi M, Galizia E, Chiorrini S, Giampieri R, Berardi R, Pierantoni C, Cascinu S: Arterial hypertension correlates with clinical outcome in colorectal cancer patients treated with first-line bevacizumab. Ann Oncol. 2009, 20: 227-230.
Schneider BP, Wang M, Radovich M, Sledge GW, Badve S, Thor A, Flockhart DA, Hancock B, Davidson N, Gralow J, Dickler M, Perez EA, Cobleigh M, Shenkier T, Edgerton S, Miller KD: Association of vascular endothelial growth factor and vascular endothelial growth factor receptor-2 genetic polymorphisms with outcome in a trial of paclitaxel compared with paclitaxel plus bevacizumab in advanced breast cancer: ECOG 2100. J Clin Oncol. 2008, 26: 4672-4678. 10.1200/JCO.2008.16.1612.
Dahlberg SE, Sandler AB, Brahmer JR, Schiller JH, Johnson DH: Clinical course of advanced non-small-cell lung cancer patients experiencing hypertension during treatment with bevacizumab in combination with carboplatin and paclitaxel on ECOG 4599. J Clin Oncol. 2010, 28: 949-954. 10.1200/JCO.2009.25.4482.
Hurwitz H, Douglas PS, Middleton JP, Sledge GW, Johnson DH, Reardon DA, Chen D, Rosen O: Analysis of early hypertension (HTN) and clinical outcome with bevacizumab (BV) [abstract]. J Clin Oncol. 2010, 28: 3039-
Dewdney A, Cunningham D, Barbachano Y, Chau I: Correlation of bevacizumab-induced hypertension and outcome in the BOXER study, a phase II study of capecitabine, oxaliplatin (CAPOX) plus bevacizumab as peri-operative treatment in 45 patients with poor-risk colorectal liver-only metastases unsuitable for upfront resection. Br J Cancer. 2012, 106 (11): 1718-1721. 10.1038/bjc.2012.152.
Bernaards C, Hegde P, Chen D, Holmgren E, Zheng M, Jubb AM, Koeppen H, Scherer SJ, Chen DS: Circulating vascular endothelial growth factor (VEGF) as a biomarker for bevacizumab-based therapy in metastatic colorectal, non-small cell lung, and renal cell cancers: analysis of phase III studies. J Clin Oncol. 2010, 28: 15s(suppl; abstr 10519)-
Duda DG, Willett CG, Ancukiewicz M, di Tomaso E, Shah M, Czito BG, Bentley R, Poleski M, Lauwers GY, Carroll M, Tyler D, Mantyh C, Shellito P, Clark JW, Jain RK: Plasma soluble VEGFR-1 is a potential dual biomarker of response and toxicity for bevacizumab with chemoradiation in locally advanced rectal cancer. Oncologist. 2010, 15: 577-583. 10.1634/theoncologist.2010-0029.
Jubb AM, Harris AL: Biomarkers to predict the clinical efficacy of bevacizumab in cancer. Lancet Oncol. 2010, 11: 1172-1183. 10.1016/S1470-2045(10)70232-1.
Wilson PM, Labonte MJ, Lenz HJ: Assessing the in vivo efficacy of biologic antiangiogenic therapies. Cancer Chemother Pharmacol. 2012, 12 (6): 468-475.
Abajo A, Boni V, Lopez I, Gonzalez-Huarriz M, Bitarte N, Rodriguez J, Zarate R, Bandres E, Garcia-Foncillas J: Identification of predictive circulating biomarkers of bevacizumab-containing regimen efficacy in pre-treated metastatic colorectal cancer patients. Br J Cancer. 2012, 107 (2): 287-290. 10.1038/bjc.2012.242.
Simkens LH, Tol J, Terstappen LW, Teerenstra S, Punt CJ, Nagtegaal ID: The predictive and prognostic value of circulating endothelial cells in advanced colorectal cancer patients receiving first-line chemotherapy and bevacizumab. Ann Oncol. 2010, 21 (12): 2447-2448. 10.1093/annonc/mdq640.
Ronzoni M, Manzoni M, Mariucci S, Loupakis F, Brugnatelli S, Bencardino K, Rovati B, Tinelli C, Falcone A, Villa E, Danova M: Circulating endothelial cells and endothelial progenitors as predictive markers of clinical response to bevacizumab-based first-line treatment in advanced colorectal cancer patients. Ann Oncol. 2010, 21 (12): 2382-2389. 10.1093/annonc/mdq261.
Matsusaka S, Suenaga M, Mishima Y, Takagi K, Terui Y, Mizunuma N, Hatake K: Circulating endothelial cells predict for response to bevacizumab-based chemotherapy in metastatic colorectal cancer. Cancer Chemother Pharmacol. 2011, 68 (3): 763-768. 10.1007/s00280-010-1543-2.
Matsusaka S, Mishima Y, Suenaga M, Terui Y, Kuniyoshi R, Mizunuma N, Hatake K: Circulating endothelial progenitors and CXCR4-positive circulating endothelial cells are predictive markers for bevacizumab. Cancer. 2011, 117 (17): 4026-4032. 10.1002/cncr.25977.
Malka D, Boige V, Jacques N, Vimond N, Adenis A, Boucher E, Pierga JY, Conroy T, Chauffert B, François E, Guichard P, Galais MP, Cvitkovic F, Ducreux M, Farace F: Clinical value of circulating endothelial cell levels in metastatic colorectal cancer patients treated with first-line chemotherapy and bevacizumab. Ann Oncol. 2012, 23 (4): 919-927. 10.1093/annonc/mdr365.
Lieb W, Safa R, Benjamin EJ, Xanthakis V, Yin X, Sullivan LM, Larson MG, Smith HM, Vita JA, Mitchell GF, Sawyer DB, Vasan RS: Vascular endothelial growth factor, its soluble receptor, and hepatocyte growth factor: clinical and genetic correlates and association with vascular function. Eur Heart J. 2009, 30: 1121-1127. 10.1093/eurheartj/ehp007.
Koutras AK, Antonacopoulou AG, Eleftheraki AG, Dimitrakopoulos FI, Koumarianou A, Varthalitis I, Fostira F, Sgouros J, Briasoulis E, Bournakis E, Bafaloukos D, Bompolaki I, Galani E, Kalogeras KT, Pectasides D, Fountzilas G, Kalofonos HP: Vascular endothelial growth factor polymorphisms and clinical outcome in colorectal cancer patients treated with irinotecan-based chemotherapy and bevacizumab. Pharmacogenomics J. in press
Lambrechts D, Claes B, Delmar P, Reumers J, Mazzone M, Yesilyurt BT, Devlieger R, Verslype C, Tejpar S, Wildiers H, de Haas S, Carmeliet P, Scherer SJ, Van Cutsem E: VEGF pathway genetic variants as biomarkers of treatment outcome with bevacizumab: an analysis of data from the AViTA and AVOREN randomised trials. Lancet Oncol. 2012, 13 (7): 724-733. 10.1016/S1470-2045(12)70231-0.
Davies RL, Grosse VA, Kucherlapati R, Bothwell M: Genetic analysis of epidermal growth factor action: assignment of human epidermal growth factor receptor gene to chromosome 7. Proc Natl Acad Sci USA. 1980, 77 (7): 4188-4192. 10.1073/pnas.77.7.4188.
Escudier B, Eisen T, Stadler WM, Szczylik C, Oudard S, Siebels M, Negrier S, Chevreau C, Solska E, Desai AA, Rolland F, Demkow T, Hutson TE, Gore M, Freeman S, Schwartz B, Shan M, Simantov R, Bukowski RM, TARGET Study Group: Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med. 2007, 356 (2): 125-134. 10.1056/NEJMoa060655.
Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Rixe O, Oudard S, Negrier S, Szczylik C, Kim ST, Chen I, Bycott PW, Baum CM, Figlin RA: Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N Engl J Med. 2007, 356 (2): 115-124. 10.1056/NEJMoa065044.
Ebi H, Corcoran RB, Singh A, Chen Z, Song Y, Lifshits E, Ryan DP, Meyerhardt JA, Benes C, Settleman J, Wong KK, Cantley LC, Engelman JA: Receptor tyrosine kinases exert dominant control over PI3K signaling in human KRAS mutant colorectal cancers. J Clin Invest. 2011, 121 (11): 4311-4321. 10.1172/JCI57909.
Saltz LB, Meropol NJ, Loehrer PJ, Needle MN, Kopit J, Mayer RJ: Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J Clin Oncol. 2004, 22 (7): 1201-1208. 10.1200/JCO.2004.10.182.
Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype C, Chau I, Van Cutsem E: Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004, 351 (4): 337-345. 10.1056/NEJMoa033025.
Vincenzi B, Santini D, Rabitti C, Coppola R, Beomonte Zobel B, Trodella L, Tonini G: Cetuximab and irinotecan as third-line therapy in advanced colorectal cancer patients: a single-centre phase II trial. Br J Cancer. 2006, 94 (6): 792-797. 10.1038/sj.bjc.6603018.
Lenz HJ, Van Cutsem E, Khambata-Ford S, Mayer RJ, Gold P, Stella P, Mirtsching B, Cohn AL, Pippas AW, Azarnia N, Tsuchihashi Z, Mauro DJ, Rowinsky EK: Multicentre phase II and translational study of cetuximab in metastatic colorectal carcinoma refractory to irintecan, oxaliplatin, and fluoropyrimidines. J Clin Oncol. 2006, 24: 4914-4921. 10.1200/JCO.2006.06.7595.
Jonker DJ, O'Callaghan CJ, Karapetis CS, Zalcberg JR, Tu D, Au HJ, Berry SR, Krahn M, Price T, Simes RJ, Tebbutt NC, van Hazel G, Wierzbicki R, Langer C, Moore MJ: Cetuximab for the treatment of colorectal cancer. N Engl J Med. 2007, 357 (20): 2040-2048. 10.1056/NEJMoa071834.
Gibson TB, Ranganathan A, Grothey A: Randomized phase III trial results of panitumumab, a fully human anti-epidermal growth factor receptor monoclonal antibody, in metastatic colorectal cancer. Clin Colorectal Cancer. 2006, 6 (1): 29-31. 10.3816/CCC.2006.n.01.
Wainberg Z, Hecht JR: A phase III randomized, open-label, controlled trial of chemotherapy and bevacizumab with or without panitumumab in the first-line treatment of patients with metastatic colorectal cancer. Clin Colorectal Cancer. 2006, 5 (5): 363-367. 10.3816/CCC.2006.n.008.
Van Cutsem E, Peeters M, Siena S, Humblet Y, Hendlisz A, Neyns B, Canon JL, Van Laethem JL, Maurel J, Richardson G, Wolf M, Amado RG: Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J Clin Oncol. 2007, 25 (13): 1658-1664. 10.1200/JCO.2006.08.1620.
Siena S, Peeters M, Van Cutsem E, Humblet Y, Conte P, Bajetta E, Comandini D, Bodoky G, Van Hazel G, Salek T, Wolf M, Devercelli G, Woolley M, Amado RG: Association of progression-free survival with patient-reported outcomes and survival: results from a randomised phase 3 trial of panitumumab. Br J Cancer. 2007, 97 (11): 1469-1474. 10.1038/sj.bjc.6604053.
Li T, Perez-Soler R: Skin toxicities associated with epidermal growth factor receptor inhibitors. Target Oncol. 2009, 4 (2): 107-119. 10.1007/s11523-009-0114-0.
Mitchell EP, Piperdi B, Lacouture ME, Shearer H, Iannotti N, Pillai MV, Xu F, Yassine M: The efficacy and safety of panitumumab administered concomitantly with FOLFIRI or irinotecan in second-line therapy for metastatic colorectal cancer: the secondary analysis from STEPP (Skin Toxicity Evaluation Protocol With Panitumumab) by KRAS status. Clin Colorectal Cancer. 2011, 10 (4): 333-339. 10.1016/j.clcc.2011.06.004.
Pinto C, Barone CA, Girolomoni G, Russi EG, Merlano MC, Ferrari D, Maiello E, American Society of Clinical Oncology, European Society of Medical Oncology: Management of skin toxicity associated with cetuximab treatment in combination with chemotherapy or radiotherapy. Oncologist. 2011, 16 (2): 228-238. 10.1634/theoncologist.2010-0298.
George TJ, Laplant KD, Walden EO, Davis AB, Riggs CE, Close JL, George SN, Lynch JW: Managing cetuximab hypersensitivity-infusion reactions: incidence, risk factors, prevention, and retreatment. J Support Oncol. 2010, 8 (2): 72-77.
Berlin J, Van Cutsem E, Peeters M, Hecht JR, Ruiz R, Wolf M, Amado RG, Meropol NJ: Predictive value of skin toxicity severity for response to panitumumab in patients with metastatic colorectal cancer (mCRC): a pooled analysis of five clinical trials [abstract]. J Clin Oncol. 2007, 25 (18S): 4134-
Stintzing S, Kapaun C, Laubender RP, Jung A, Neumann J, Modest DP, Giessen C, Moosmann N, Wollenberg A, Kirchner T, Heinemann V: Prognostic value of cetuximab related skin toxicity in metastatic colorectal cancer (mCRC) patients and its correlation with parameters of the EGFR signal transduction pathway. Results from a randomized trial of the GERMAN AIO CRC Study Group. Int J Cancer. 2013, 132 (1): 236-245. 10.1002/ijc.27654.
Saif MW, Kaley K, Chu E, Copur MS: Safety and efficacy of panitumumab therapy after progression with cetuximab: experience at two institutions. Clin Colorectal Cancer. 2010, 9 (5): 315-318. 10.3816/CCC.2010.n.046.
Peeters M, Siena S, Van Cutsem E, Sobrero A, Hendlisz A, Cascinu S, Kalofonos H, Devercelli G, Wolf M, Amado RG: Association of progression-free survival, overall survival, and patient-reported outcomes by skin toxicity and KRAS status in patients receiving panitumumab monotherapy. Cancer. 2009, 115 (7): 1544-1554. 10.1002/cncr.24088.
Lacouture ME, Mitchell EP, Shearer H, Iannotti N, Piperdi B, Pillai MV, Xu F, Yassine M: Impact of pre-emptive skin toxicity treatment on panitumumab-related skin toxicities quality of life in patients with metastatic colorectal cancer: Results from STEPP. ASCO Gastrointestinal Cancers Symposium: 15–17 January. 2009, San Francisco, CA. Alexandria: ASCO
Graziano F, Ruzzo A, Loupakis F, Canestrari E, Santini D, Catalano V, Bisonni R, Torresi U, Floriani I, Schiavon G, Andreoni F, Maltese P, Rulli E, Humar B, Falcone A, Giustini L, Tonini G, Fontana A, Masi G, Magnani M: Pharmacogenetic profiling for cetuximab plus irinotecan therapy in patients with refractory advanced colorectal cancer. J Clin Oncol. 2008, 26 (9): 1427-1434. 10.1200/JCO.2007.12.4602.
Garm Spindler KL, Pallisgaard N, Rasmussen AA, Lindebjerg J, Andersen RF, Crüger D, Jakobsen A: The importance of KRAS mutations and EGF61A>G polymorphism to the effect of cetuximab and irinotecan in metastatic colorectal cancer. Ann Oncol. 2009, 20 (5): 879-884. 10.1093/annonc/mdn712.
Mok T, Wu YL, Zhang L: A small step towards personalized medicine for non-small cell lung cancer. Discov Med. 2009, 8 (43): 227-231.
Pander J, Gelderblom H, Antonini NF, Tol J, van Krieken JH, van der Straaten T, Punt CJ, Guchelaar HJ: Correlation of FCGR3A and EGFR germline polymorphisms with the efficacy of cetuximab in KRAS wild-type metastatic colorectal cancer. Eur J Cancer. 2010, 46 (10): 1829-1834. 10.1016/j.ejca.2010.03.017.
Dahan L, Norguet E, Etienne-Grimaldi MC, Formento JL, Gasmi M, Nanni I, Gaudart J, Garcia S, Ouafik L, Seitz JF, Milano G: Pharmacogenetic profiling and cetuximab outcome in patients with advanced colorectal cancer. BMC Cancer. 2011, 11: 496-10.1186/1471-2407-11-496.
Grossman AH, Samowitz WS: Epidermal growth factor receptor pathway mutations and colorectal cancer therapy. Arch Pathol Lab Med. 2011, 135: 1278-1282. 10.5858/arpa.2011-0047-RA.
Ålgars A, Lintunen M, Carpén O, Ristamäki R, Sundström J: EGFR gene copy number assessment from areas with highest EGFR expression predicts response to anti-EGFR therapy in colorectal cancer. Br J Cancer. 2011, 105 (2): 255-262. 10.1038/bjc.2011.223.
Ensinger C, Sterlacci W: Implications of EGFR PharmDx kit for cetuximab eligibility. Expert Rev Mol Diagn. 2008, 8 (2): 141-148. 10.1586/14737159.8.2.141.
Chung KY, Shia J, Kemeny NE, Shah M, Schwartz GK, Tse A, Hamilton A, Pan D, Schrag D, Schwartz L, Klimstra DS, Fridman D, Kelsen DP, Saltz LB: Cetuximab shows activity in CRC patients with tumours that do not express the EGFR by immunohistochemistry. J Clin Oncol. 2005, 23: 1803-1810. 10.1200/JCO.2005.08.037.
Kang MJ, Hong YS, Kim KP, Kim SY, Baek JY, Ryu MH, Lee JL, Chang HM, Kim MJ, Chang HJ, Kang YK, Kim TW: Biweekly cetuximab plus irinotecan as second-line chemotherapy for patients with irinotecan-refractory and KRAS wild-type metastatic colorectal cancer according to epidermal growth factor receptor expression status. Invest New Drugs. 2011, 30 (4): 1607-1613.
Fearon ER: Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011, 6: 479-507. 10.1146/annurev-pathol-011110-130235.
Souglakos J, Philips J, Wang R, Marwah S, Silver M, Tzardi M, Silver J, Ogino S, Hooshmand S, Kwak E, Freed E, Meyerhardt JA, Saridaki Z, Georgoulias V, Finkelstein D, Fuchs CS, Kulke MH, Shivdasani RA: Prognostic and predictive value of common mutations for treatment response and survival in patients with metastatic colorectal cancer. Br J Cancer. 2009, 101 (3): 465-472. 10.1038/sj.bjc.6605164.
Andreyev HJ, Tilsed JV, Cunningham D, Sampson SA, Norman AR, Schneider HJ, Clarke PA: K-ras mutations in patients with early colorectal cancers. Gut. 1997, 41 (3): 323-329. 10.1136/gut.41.3.323.
Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA: Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. J Natl Cancer Inst. 1998, 90 (9): 675-684. 10.1093/jnci/90.9.675.
Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, Young J, Walsh T, Ward R, Hawkins N, Beranek M, Jandik P, Benamouzig R, Jullian E, Laurent-Puig P, Olschwang S, Muller O, Hoffmann I, Rabes HM, Zietz C, Troungos C, Valavanis C, Yuen ST, Ho JW, Croke CT, O'Donoghue DP, Giaretti W, Rapallo A, Russo A, Bazan V: Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br J Cancer. 2011, 85 (5): 692-696.
Edkins S, O'Meara S, Parker A, Stevens C, Reis M, Jones S, Greenman C, Davies H, Dalgliesh G, Forbes S, Hunter C, Smith R, Stephens P, Goldstraw P, Nicholson A, Chan TL, Velculescu VE, Yuen ST, Leung SY, Stratton MR, Futreal PA: Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol Ther. 2006, 5 (8): 928-932. 10.4161/cbt.5.8.3251.
Janakiraman M, Vakiani E, Zeng Z, Pratilas CA, Taylor BS, Chitale D, Halilovic E, Wilson M, Huberman K, Ricarte Filho JC, Persaud Y, Levine DA, Fagin JA, Jhanwar SC, Mariadason JM, Lash A, Ladanyi M, Saltz LB, Heguy A, Paty PB, Solit DB: Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res. 2010, 70 (14): 5901-5911. 10.1158/0008-5472.CAN-10-0192.
Lièvre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Côté JF, Tomasic G, Penna C, Ducreux M, Rougier P, Penault-Llorca F, Laurent-Puig P: KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006, 66 (8): 3992-3995. 10.1158/0008-5472.CAN-06-0191.
Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD: Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008, 26 (10): 1626-1634. 10.1200/JCO.2007.14.7116.
Lin JS, Webber EM, Senger CA, Holmes RS, Whitlock EP: Systematic review of pharmacogenetic testing for predicting clinical benefit to anti-EGFR therapy in met CRC. Am J Cancer Res. 2011, 1 (5): 650-662.
Loupakis F, Cremolini C, Salvatore L, Schirripa M, Lonardi S, Vaccaro V, Cuppone F, Giannarelli D, Zagonel V, Cognetti F, Tortora G, Falcone A, Bria E: Clinical impact of anti-epidermal growth factor receptor monoclonal antibodies in first-line treatment of metastatic colorectal cancer: meta-analytical estimation and implications for therapeutic strategies. Cancer. 2012, 118 (6): 1523-1532. 10.1002/cncr.26460.
Vale CL, Tierney JF, Fisher D, Adams RA, Kaplan R, Maughan TS, Parmar MK, Meade AM: Does anti-EGFR therapy improve outcome in advanced colorectal cancer? A systematic review and meta-analysis. Cancer Treat Rev. 2012, 38 (6): 618-625. 10.1016/j.ctrv.2011.11.002.
Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, McAllister PK, Morton RF, Schilsky RL: American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009, 27 (12): 2091-2096. 10.1200/JCO.2009.21.9170.
Cripps C, Gill S, Ahmed S, Colwell B, Dowden S, Kennecke H, Maroun J, Samson B, Thirlwell M, Wong R: Consensus recommendations for the use of anti-EGFR therapies in metastatic colorectal cancer. Curr Oncol. 2010, 17 (6): 39-45.
Aubin F, Gill S, Burkes R, Colwell B, Kamel-Reid S, Koski S, Pollett A, Samson B, Tehfe M, Wong R, Young S, Soulières D: Canadian Expert Group consensus recommendations: KRAS testing in colorectal cancer. Curr Oncol. 2011, 8 (4): e180-e184.
De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, Penault-Llorca F, Rougier P, Vincenzi B, Santini D, Tonini G, Cappuzzo F, Frattini M, Molinari F, Saletti P, De Dosso S, Martini M, Bardelli A, Siena S, Sartore-Bianchi A, Tabernero J, Macarulla T, Di Fiore F, Gangloff AO, Ciardiello F, Pfeiffer P: Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010, 11 (8): 753-762. 10.1016/S1470-2045(10)70130-3.
Baldus SE, Schaefer KL, Engers R, Hartleb D, Stoecklein NH, Gabbert HE: Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res. 2010, 16 (3): 790-799. 10.1158/1078-0432.CCR-09-2446.
Mariani P, Lae M, Degeorges A, Cacheux W, Lappartient E, Margogne A, Pierga JY, Girre V, Mignot L, Falcou MC, Salmon RJ, Delattre O, De Cremoux P: Concordant analysis of KRAS status in primary colon carcinoma and matched metastasis. Anticancer Res. 2010, 30 (10): 4229-4235.
Bösmüller H, Kranewitter W, Webersinke G, Rumpold H, Hackl M, Fend F: Morphological and molecular heterogeneity in colorectal neoplasms with K-RAS mutation. A report of two cases. Pathol Res Pract. 2011, 207 (6): 399-402. 10.1016/j.prp.2011.03.004.
Farber L, Efrati E, Elkin H, Peerless Y, Sabo E, Ben-Izhak O, Hershkovitz D: Molecular morphometric analysis shows relative intra-tumoural homogeneity for KRAS mutations in colorectal cancer. Virchows Arch. 2011, 459 (5): 487-493. 10.1007/s00428-011-1158-y.
Oltedal S, Aasprong OG, Møller JH, Kørner H, Gilje B, Tjensvoll K, Birkemeyer EM, Heikkilä R, Smaaland R, Nordgård O: Heterogeneous distribution of K-ras mutations in primary colon carcinomas: implications for EGFR-directed therapy. Int J Colorectal Dis. 2011, 26 (10): 1271-1277. 10.1007/s00384-011-1233-5.
Franklin WA, Haney J, Sugita M, Bemis L, Jimeno A, Messersmith WA: KRAS mutation: comparison of testing methods and tissue sampling techniques in colon cancer. J Mol Diagn. 2010, 12 (1): 43-50. 10.2353/jmoldx.2010.080131.
Richman SD, Chambers P, Seymour MT, Daly C, Grant S, Hemmings G, Quirke P: Intra-tumoral heterogeneity of KRAS and BRAF mutation status in patients with advanced colorectal cancer (aCRC) and cost effectiveness of multiple sample testing. Anal Cell Pathol (Amst). 2011, 34 (1–2): 61-66.
Blank PR, Moch H, Szucs TD, Schwenkglenks M: KRAS and BRAF mutation analysis in metastatic colorectal cancer: a cost-effectiveness analysis from a Swiss perspective. Clin Cancer Res. 2011, 17 (19): 6338-6346. 10.1158/1078-0432.CCR-10-2267.
Vijayaraghavan A, Efrusy MB, Göke B, Kirchner T, Santas CC, Goldberg RM: Cost-effectiveness of KRAS testing in metastatic colorectal cancer patients in the United States and Germany. Int J Cancer. 2012, 131 (2): 438-445. 10.1002/ijc.26400.
French AJ, Sargent DJ, Burgart LJ, Foster NR, Kabat BF, Goldberg R, Shepherd L, Windschitl HE, Thibodeau SN: Prognostic significance of defective mismatch repair and BRAF V600E in patients with colon cancer. Clin Cancer Res. 2008, 14 (11): 3408-3415. 10.1158/1078-0432.CCR-07-1489.
Laurent-Puig P, Cayre A, Manceau G, Buc E, Bachet JB, Lecomte T, Rougier P, Lievre A, Landi B, Boige V, Ducreux M, Ychou M, Bibeau F, Bouché O, Reid J, Stone S, Penault-Llorca F: Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol. 2009, 27 (35): 5924-5930. 10.1200/JCO.2008.21.6796.
Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A: Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008, 26 (35): 5705-5712. 10.1200/JCO.2008.18.0786.
Mao C, Liao RY, Qiu LX, Wang XW, Ding H, Chen Q: BRAF V600E mutation and resistance to anti-EGFR monoclonal antibodies in patients with metastatic colorectal cancer: a meta-analysis. Mol Biol Rep. 2011, 38 (4): 2219-2223. 10.1007/s11033-010-0351-4.
Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, Beijersbergen RL, Bardelli A, Bernards R: Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012, 483 (7387): 100-103.
Zhao L, Vogt PK: Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci USA. 2008, 105 (7): 2652-2657. 10.1073/pnas.0712169105.
Kennedy AL, Morton JP, Manoharan I, Nelson DM, Jamieson NB, Pawlikowski JS, McBryan T, Doyle B, McKay C, Oien KA, Enders GH, Zhang R, Sansom OJ, Adams PD: Activation of the PIK3CA/AKT pathway suppresses senescence induced by an activated RAS oncogene to promote tumorigenesis. Mol Cell. 2011, 42 (1): 36-49. 10.1016/j.molcel.2011.02.020.
Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, Di Nicolantonio F, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A: PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009, 69 (5): 1851-1857. 10.1158/0008-5472.CAN-08-2466.
Prenen H, De Schutter J, Jacobs B, De Roock W, Biesmans B, Claes B, Lambrechts D, Van Cutsem E, Tejpar S: PIK3CA mutations are not a major determinant of resistance to the epidermal growth factor receptor inhibitor cetuximab in metastatic colorectal cancer. Clin Cancer Res. 2009, 15 (9): 3184-3188. 10.1158/1078-0432.CCR-08-2961.
Goel A, Arnold CN, Niedzwiecki D, Carethers JM, Dowell JM, Wasserman L, Compton C, Mayer RJ, Bertagnolli MM, Boland CR: Frequent inactivation of PTEN by promoter hypermethylation in microsatellite instability-high sporadic colorectal cancers. Cancer Res. 2004, 64 (9): 3014-3021. 10.1158/0008-5472.CAN-2401-2.
Sartore-Bianchi A, Di Nicolantonio F, Nichelatti M, Molinari F, De Dosso S, Saletti P, Martini M, Cipani T, Marrapese G, Mazzucchelli L, Lamba S, Veronese S, Frattini M, Bardelli A, Siena S: Multi-determinants analysis of molecular alterations for predicting clinical benefit to EGFR-targeted monoclonal antibodies in colorectal cancer. PLoS One. 2009, 4 (10): e7287-10.1371/journal.pone.0007287.
Perrone F, Lampis A, Orsenigo M, Di Bartolomeo M, Gevorgyan A, Losa M, Frattini M, Riva C, Andreola S, Bajetta E, Bertario L, Leo E, Pierotti MA, Pilotti S: PI3KCA/PTEN deregulation contributes to impaired responses to cetuximab in metastatic colorectal cancer patients. Ann Oncol. 2009, 20 (1): 84-90.
Sood A, McClain D, Maitra R, Basu-Mallick A, Seetharam R, Kaubisch A, Rajdev L, Mariadason JM, Tanaka K, Goel S: PTEN gene expression and mutations in the PIK3CA gene as predictors of clinical benefit to anti-epidermal growth factor receptor antibody therapy in patients with KRAS wild-type metastatic colorectal cancer. Clin Colorectal Cancer. 2012, 11 (2): 143-150. 10.1016/j.clcc.2011.12.001.
Ulivi P, Capelli L, Valgiusti M, Zoli W, Scarpi E, Chiadini E, Rosetti P, Bravaccini S, Calistri D, Saragoni L, Casadei Gardini A, Ragazzini A, Frassineti GL, Amadori D, Passardi A: Predictive role of multiple gene alterations in response to cetuximab in metastatic colorectal cancer: a single center study. J Transl Med. 2012, 10 (1): 87-10.1186/1479-5876-10-87.
Frattini M, Saletti P, Romagnani E, Martin V, Molinari F, Ghisletta M, Camponovo A, Etienne LL, Cavalli F, Mazzucchelli L: PTEN loss of expression predicts cetuximab efficacy in metastatic colorectal cancer patients. Br J Cancer. 2007, 97 (8): 1139-1145. 10.1038/sj.bjc.6604009.
Tol J, Dijkstra JR, Klomp M, Teerenstra S, Dommerholt M, Vink-Börger ME, van Cleef PH, van Krieken JH, Punt CJ, Nagtegaal ID: Markers for EGFR pathway activation as predictor of outcome in metastatic colorectal cancer patients treated with or without cetuximab. Eur J Cancer. 2010, 46 (11): 1997-2009. 10.1016/j.ejca.2010.03.036.
Loupakis F, Pollina L, Stasi I, Ruzzo A, Scartozzi M, Santini D, Masi G, Graziano F, Cremolini C, Rulli E, Canestrari E, Funel N, Schiavon G, Petrini I, Magnani M, Tonini G, Campani D, Floriani I, Cascinu S, Falcone A: PTEN expression and KRAS mutations on primary tumors and metastases in the prediction of benefit from cetuximab plus irinotecan for patients with metastatic colorectal cancer. J Clin Oncol. 2009, 27 (16): 2622-2629. 10.1200/JCO.2008.20.2796.
Negri FV, Bozzetti C, Lagrasta CA, Crafa P, Bonasoni MP, Camisa R, Pedrazzi G, Ardizzoni A: PTEN status in advanced colorectal cancer treated with cetuximab. Br J Cancer. 2010, 102 (1): 162-164. 10.1038/sj.bjc.6605471.
Barbier A, Domont J, Magné N, Goldmard JL, Genestie C, Hannoun C, Vaillant JC, Bellanger A, Khayat D, Capron F, Spano JP: Coexpression of biological key modulators in primary colorectal carcinomas and related metastatic sites: implications for treatment with cetuximab. Bull Cancer. 2010, 97 (2): E9-E15.
Perkins G, Lièvre A, Ramacci C, Méatchi T, de Reynies A, Emile JF, Boige V, Tomasic G, Bachet JB, Bibeau F, Bouché O, Penault-Llorca F, Merlin JL, Laurent-Puig P: Additional value of EGFR downstream signaling phosphoprotein expression to KRAS status for response to anti-EGFR antibodies in colorectal cancer. Int J Cancer. 2010, 127 (6): 1321-1331. 10.1002/ijc.25152.
FDA: Draft guidance for Industry and Food and Drug Administration Staff - in vitro companion diagnostic devices. [http://www.fda.gov/medicaldevices/deviceregulationandguidance/guidancedocuments/ucm262292.htm]
Rinaldi F, George E, Adler AI: NICE guidance on cetuximab, bevacizumab, and panitumumab for treatment of metastatic colorectal cancer after first-line chemotherapy. Lancet Oncol. 2012, 13 (3): 233-234. 10.1016/S1470-2045(12)70044-X.
Characiejus D, Hodzic J, Jacobs JJL: “First do no harm” and the importance of prediction in oncology. EPMA J. 2010, 1 (3): 369-375. 10.1007/s13167-010-0042-1.
Clausson CM, Grundberg I, Weibrecht I, Nilsson M, Söderberg O: Methods for analysis of the cancer microenvironment and their potential for disease prediction, monitoring and personalized treatments. EPMA J. 2012, 3: 7-
Ibrahim AEK, Arends MJ: Molecular typing of colorectal cancer: applications in diagnosis and treatment. Diagnostic Histopathology. 2012, 18 (2): 70-80. 10.1016/j.mpdhp.2011.11.002.
Acknowledgements
SH is employed by the Scottish Higher Education Funding Council, UK. MO is an employee and shareholder of AstraZeneca, Cheshire, UK. BD is funded by the Health Research Board (PDTM/2011/18).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SH and BD conceived the review, and SH coordinated the drafting of the manuscript. SH, MO and BD participated in the design of the review, performed literature searches and identified relevant studies. BD and MO provided content expertise. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Hagan, S., Orr, M.C.M. & Doyle, B. Targeted therapies in colorectal cancer—an integrative view by PPPM. EPMA Journal 4, 3 (2013). https://doi.org/10.1186/1878-5085-4-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1878-5085-4-3