Abstract
Background
We have previously demonstrated that phenylhexyl isothiocyanate (PHI), a synthetic isothiocyanate, inhibits histone deacetylases and remodels chromatins to induce growth arrest in HL-60 myeloid leukemia cells in a concentration-dependent manner.
Methods
To investigate the effect of PHI, a novel histone deacetylases inhibitor (HDACi), on demethylation and activation of transcription of p15 in acute lymphoid leukemia cell line Molt-4, and to further decipher the potential mechanism of demethylation, DNA sequencing and modified methylation specific PCR (MSP) were used to screen p15-M and p15-U mRNA after Molt-4 cells were treated with PHI, 5-Aza and TSA. DNA methyltransferase 1 (DNMT1), 3A (DNMT3A), 3B (DNMT3B) and p15 mRNA were measured by RT-PCR. P15 protein, acetylated histone H3 and histone H4 were detected by Western Blot.
Results
The gene p15 in Molt-4 cells was hypermethylated and inactive. Hypermethylation of gene p15 was attenuated and p15 gene was activated de novo after 5 days exposure to PHI in a concentration-dependent manner. DNMT1 and DNMT3B were inhibited by PHI (P < 0.05). Alteration of DNMT3A was not significant at those concentrations. Acetylated histone H3 and histone H4 were accumulated markedly after exposure to PHI.
Conclusion
PHI could induce both DNA demethylation and acetylated H3 and H4 accumulation in Molt-4 cells. Hypermethylation of gene p15 was reversed and p15 transcription could be reactivated de novo by PHI.
Similar content being viewed by others
Background
The major epigenetic transcriptional controls involved in gene silencing are DNA methylation and covalent modification of histone proteins. Transcriptional silencing of genes, due to hypermethylation of CpG islands in the promoter region of genes, has been reported in nearly every type of human tumors. A broad spectrum of genes are frequently hypermethylated in cancers, including those associated with cell cycle regulation, detoxification, tumor suppression, and apoptosis etc. DNA methylation is catalyzed by DNA methyltransferases (DNMTs), of which three active enzymes have been identified in mammals, namely DNMT1, DNMT3A and DNMT3B. In developmental processes of the mouse, DNMT1 is responsible for maintaining pre-existing methylation patterns during DNA replication, while DNMT3A and DNMT3B are required for initiation of de novo methylation. DNMT3A expression is ubiquitous, but DNMT3B is present in the cells at low levels except in testes, thyroid, and bone marrow[1]. DNMTs play roles in gene silencing by acting as transcriptional repressors themselves, or by serving as binding scaffolds for transcriptional repressors, histone deacetylases and histone methyltransferases. Thereby, DNMTs can establish gene silencing independent of their catalytic activities[2–4]. In human leukemia cells, the DNMTs are found to be aberrantly over-expressed[5, 6]. Histone proteins assemble into nucleosomes, which function as both DNA packaging units and transcriptional regulators. The amino-terminal tails of histones protrude from the nucleosome and they are subjected to covalent modifications such as acetylation, methylation and phosphorylation. The different types of histone modifications have been linked with distinct functions. Modifications to histones influence chromatin structure, and ultimately gene transcription, including those coding for tumor suppressor proteins. One of the key histone modification that control gene transcription is acetylation, which is regulated by two opposing enzymatic activities (histone acetyltransferases [HATs] and histone deacetylases [HDACs])[7]. HATs are in charge of histone acetylation, leading to the relaxation of chromatin structure and transcriptional activation of genes, while HDACs are in charge of histone deacetylation, which is associated generally with chromatin condensation and transcriptional repression[8].
The gene p15 (INK4b or MTS2) is a candidate tumor suppressor with structural and functional similarity to the p16 gene. This gene recognizes cyclin-dependent kinases CDK4 and CDK6, and induces G1 arrest of the cell cycle by competing with cyclin D for binding with CDK4. Specific deletions of p15 sequences have been found in only a few cases of leukemia and lymphomas. In contrast, the p15 gene is preferentially hypermethylated at a 5'-CpG island, which has been shown to be associated with loss of transcription of this gene in leukemia cells [9, 10]. Furthermore, aberrant p15 methylation seems to have important prognostic implications for risk assessment because patients with p15 methylation have overall shortened survival [11, 12].
We have previously demonstrated that phenylhexyl isothiocyanate (PHI), a man-made isothiocyanate, inhibits histone deacetylases and remodels chromatins to induce growth arrest in HL-60 myeloid leukemia cells in a concentration-dependent manner[13]. Recent research has described that this class of small chemicals, either present naturally in cruciferous vegetables or man-made are potential chemopreventive agents. In animal models, PHI was effective against tumorigenesis of esophagus, leukemia, and carcinomas of lung and prostate[14–17]. The major mechanism includes the induction of growth arrest and apoptosis in tumor cells[18, 19]. Since we have demonstrated that PHI is an inhibitor of HDACs, its effects on p15 activity in leukemia cells has been a subject of investigation. In this paper we demonstrated that PHI has a dual effect on DNA methylation and histone acetylation in leukemic T cells Molt-4. The cross-talk on the DNA and chromatin resulted in demethylating the CpG island of p15, which is inactivated due to hypermethylation, and recovered the unmethylated p15 for transcriptional activation. An inter-active mechanism, involving a down-regulation of the enzymes DNMTs and up-regulating the histone acetyltransferase P300/CBP and histone acetylation, is revealed.
Methods
Cell cultures
PHI, greater than 98% pure, was purchased from LKT Lab (St. Paul, MN). Human acute lymphatic leukemia cell line Molt-4 was obtained from China Center for Type Culture Collection (CCTCC). Cells were maintained in RPMI-1640 medium supplemented with 10% heat-inactivated fetal calf serum and maintained at 37°C in humidified atmosphere containing 5% CO2. Cells in exponential growth were exposed to PHI prepared in 75% methanol[13] at various concentrations for 5 days. The control cultures were supplemented with the methanol-containing medium. Some cell cultures were supplemented with 2 μM of 5-azacytidine (5-Aza), a known inhibitor of DNA methylation, or 1 μM of Trichostatin A (TSA), a known inhibitor of histone deacetylases, at various concentrations.
Methylation specific PCR (MSP)
The genomic DNA from cultured cells was extracted and modified by bisulfate treatment for MS-PCR analyses[20]. DNA from cell cultures under different conditions was isolated with the Tissue/Cell Genomic DNA Isolation Kit (Pearl, China), employing the Wizard DNA Clean-Up System (Promega, USA), and amplified by PCR with two sets of gene promoter specific primer pairs that recognize the methylated (M) and the unmethylated (U) CpG sites. The primers for the methylated form of p15 (148pb) were gcgttcgtattttgcggtt (positive sense), and cgtacaataaccgaacgaccga (antisense). The primers for unmethylated form (154 bp) were tgtgatgtgtttgtattttgtggtt (positive sense) and ccatacaataaccaaacaaccaa (antisense). The amplification was performed in an Mastercycler unit ( Eppendorf) under the program conditions as follows: 95°C for 5 min; then 40 cycles of 95°C for 45 sec, 60°C for 45 sec, 72°C for 45 sec; and finally 10 min at 72°C. The PCR products were visualized in GeneGenius (Syngene, British) by ethidium bromide staining in 2% agarose gels.
Western blot analysis
The protein levels were determined by Western blot analysis as described previously[21]. Briefly, total proteins were prepared from each culture condition with a lysis buffer containing protease inhibitors, and the lysates collected after centrifugation at 4°C. The protein content of the lysates was determined with the Bradford protein assay. Protein lysate was subjected to SDS-PAGE, electrotransferred to nitrocellulose membrane, and immunoblotted with specific antibodies. The following antibodies were used for immunoblotting: anti-acetyl-histone H3 (lysines 9 and 14) (1:300 dilution, Upstate), anti-acetyl-Histone H4 (lysines 5, 8, 12, and 16) (1:300 dilution, Upstate), and anti-300/CBP (1:300 dilution, Upstate). Antibodies against P15, and a goat anti-rabbit with HRP conjugate secondary antibodies were purchased from Santa Cruz (USA). The reaction was visualized through the ECL system, and the protein levels were quantified with the anti-β-actin protein estimation assay.
Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
Total RNA was isolated using Trizol reagent (Invitrogen, USA) according to the manufacturer's instructions. One-microgram total RNA isolated was used for the first-strand cDNA synthesis with Reverse Transcription System (Promega, USA). cDNA was amplified using specific primer for p15, DNMT1, DNMT3A, DNMT3B in separate reactions (Table 1). β-actin was used as a loading control to ensure that cDNA was complete and Taq was deactivated in each reaction. The PCR products were visualized in GeneGenius (Syngene, British) by ethidium bromide staining in 1.6% agarose gels.
Statistical analysis
The data was analyzed by statistical software SPSS13.0. Data measurement was presented as mean ± SD, from multiple independent experiments by homogeneity test for variance and test of normality. Results were evaluated by One-way ANOVA between groups. P < 0.05 was considered to be statistically significant.
Results
Demethylating p15 by PHI
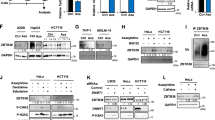
The DNA methylation status of p15 in human lymphatic leukemia T cells Molt-4 was evaluated by MS-PCR. Figure 1A showed the presence of methylated p15 (p15-M), and the unmethylated p15 (p15-U) was undetectable. After exposure of Molt-4 cells to PHI for 5 days, the methylated p15 was decreased, with the magnitude in positive relation to the PHI concentrations. The unmethylated p15 became detectable, replacing the decrease of methylated p15. The activity of 40 μM PHI to reverse p15 methylation was similar to that of 2 μM of 5-Aza, a known inhibitor of DNA methylation or 1 μM of TSA, which was shown in endometrial cancer cells as a demethylating agent as it reduced DNMT3B level and de novo DNMT activity[22].
PHI reversed hypermethylation of p15 and induced transcription activation. A: PHI induced p15 hypomethylation with the decreasing of the methylated p15 (p15-M) and increase of the unmethylated form of p15 (p15-U). MS-PCR was performed using primers specific for p15-M and p15-U DNA forms of p15 as described in the methods. 5-Aza (2 μM) and TSA (1 μM) were used as controls for DNA hypomethylation. B: P15 mRNA levels were enhanced by PHI treatment. The expression levels of p15 mRNA were measured by RT-PCR. C: Up-regulation of p15 protein expression by PHI. P15 protein levels were detected by Western blotting, and β-actin was used as a loading control.
The mRNA level of p15 in Molt-4 cells was examined without or with the exposure to PHI. Figure 1B showed the comparison of the levels of mRNA of p15 and β-actin. In the presence of PHI, the p15 mRNA expression was enhanced in a concentration-dependent manner. The ratios were: control (0.17 ± 0.12), PHI 10 μM (0.29 ± 0.14), PHI 20 μM (0.55 ± 0.07), PHI 40 μM (0.93 ± 0.13), TSA (0.65 ± 0.11), and 5-Aza (0.89 ± 0.13). The expression levels of p15 mRNA mediated by PHI were statistically significantly (P < 0.05) as compared to that without PHI treatment. The protein expression of p15 was examined by Western blotting in parallel. Figure 1C depicts convincingly a dose-related increase of p15 expression after treatment with PHI at 20-40 μM, similar to the 5-Aza effect. The results thus indicated that PHI might be a potent methylation inhibitor for the CpG island of p15 gene, leading to reactivating p15 transcription in the leukemic cells.
PHI decreased the expression of DNMT1, DNMT3B
The mRNA levels of the enzymes responsible for DNA methylation, i.e., DNMT1, DNMT3A, DNMT3B, without or with the exposure to PHI, were evaluated with RT-PCR (Figure 2A). The gray scale of DNMTs was contrasted to that of β-actin (Figure 2B), and demonstrated that the mRNA of DNMT1 and DNMT3B were significantly decreased after exposure for to PHI for 5 days, in a concentration-dependent manner (P < 0.05). The mRNA level of DNMT3A, on the other hand, was not significantly altered under the same condition.
PHI induced histone acetylation and acetyltransferase up-regulation
Molt-4 cells were exposed to PHI at various concentrations. Figure 3A showed that after exposure of Molt-4 cells to PHI, the acetylation of histone H3 or H4 was significantly increased in a concentration and time-dependent manner. Acetylated histone H3 was elevated approximately 1.13, 1.21 and 1.35-folds with PHI at 5, 20, or 40 μM for 3 h, as compared to controls without PHI. After 7 hours, acetylation was increased by 2.0, 2.2, 4.0-folds. Similarly, acetylated histone H4 was increased approximately 1.09, 1.45 and 1.72-folds after 3 hours, and 1.3, 1.8, 1.9-folds after 7 hours. The enzyme level of P300/CBP was examined in parallel. Figure 3B showed that the increase of P300/CBP could be clearly observed after exposed to 5 μM or more PHI, similar to the effects on histone acetylation. Approximately 1.1, 1.13 and 1.23-folds increase of P300/CBP, over the control, was observed 3 hours after exposure to PHI at 5, 20 and 40 μM, and approximately 1.38, 1.9 and 2.14-folds after 7 hours.
PHI induced histone acetylation and acetyltransferase up-regulation. A: The acetylation of histone H3 and H4 was significantly increased in a concentration and time-dependent manner by PHI. B: PHI up-regulated the expression of acetyltransferase P300/CBP in a concentration and time-dependent manner.
Discussion
DNA methylation and histone acetylation are two well known epigenetic chromatin modifications. At least 80 clinical trials are underway, testing more than eleven different HDAC inhibitory agents including both hematological and solid malignancies [23–25]. This study showed for the first time that PHI had dual effects on enhancing histone acetylation as well as demethylating p15. Upon exposure to PHI, the unmethylated p15, otherwise absent, became detectable, along with the disappearance of the methylated p15. This reversal of p15 methylation correlates with reexpression of p15. The activity of PHI in demethylating p15 was shown to be similar to that of 5-Aza and TSA. To our knowledge, this study is the first to demonstrate that the hypermethylated p15 gene in leukemic T cells could be reactivated by an isothiocyanate. Our analyses have provided two inter-related mechanisms for hypomethylation of p15. They are presented in the following.
One potential mechanism for initiating demethylation of p15 by PHI could be that the expressions of the DNA methylating enzymes were down-regulated. This possibility was examined with the three DNA methylating enzymes DNMT1, DNMT3A, and DNMT3B. The study showed that in the presence of PHI, the mRNA of two of the enzymes, DNMT1 and DNMT3B, was significantly down-regulated in a concentration-dependent manner. The mRNA of the enzyme DNMT3A, however, did not show a significant alteration. The expression of DNMT3A is known to be ubiquitous and this has been under investigation as a basis of this observation. The results suggested that reduction of the DNMT1 and DNMT3B expression and their activity could be a responsible mechanism for hypomethylation of p15. This interpretation is in line with the reports that DNMTs are commonly over-expressed in leukemias. AML cells with hypermethylated p15 tended to express higher levels of DNMT1 and DNMT3B [5, 6]. Melki et al [5] demonstrated that the average expression of DNA methyltransferase mRNA from the bone marrow cells of leukemic patients is elevated 4.4-folds, as compared to the expression of normal bone marrow. Mizuno[6] found a mean increase of 5.3-, 4.4-, and 11.7-folds of DNMT1, 3A, and 3B in AML, respectively, comparing with the control bone marrow cells. Although CML cells in the chronic phase did not show significant changes, cells in the acute phase showed 3.2-, 4.5-, and 3.4- fold mean increases in the levels of DNMT1, 3A, and 3B, respectively.
The major mechanism of the current hypomethylating drugs, such 5-Azacytidine and decitabine, is their covalent binding to the DNMTs, which resulting in the irreversible inhibition of the DNMT activity, leading to the hypomethylation of the genomic DNA [26]. Our data showed that PHI could reduce the synthesis of DNMTs. It may be an additional mechanism for PHI to induce DNA hypomethylation. Whether PHI also bind to DNMTs remains to be investigated.
The methylation of p15 by PHI could also be related to the PHI effects on histone acetylation. Histone acetylation generally correlates to an open and transcriptionally active chromatin, whereas histone deacetylation is associated with chromatin condensation and transcriptional repression. The p15 CpG island region is surrounded with both histone acetylated H3 and methylated H3K9 in AML[27]. PHI could up-regulate the expression of acetyltransferase P300/CBP and induces the accumulation of acetylated histone H3, H4 in Molt-4, resulting in chromatin unfolding and accessibility of regulators in the p15 promoter for transcriptional activation. This is consistent with previous data showing that PHI was inhibitor of HDACs [28].
Conclusion
PHI could induce DNA demethylation and acetylated H3, H4 accumulation in Molt-4 cells. Hypermethylation of gene p15 was reversed and p15 transcription could be reactivated de novo by PHI.
Abbreviations
- PHI:
-
Phenylhexyl Isothiocyanate
- HDAC:
-
histone deacetylases
- MSP:
-
Methylation Specific PCR
- 5-Aza:
-
5-azacytidine
- TSA:
-
Trichostatin A
- DNMT:
-
DNA methyltransferase
- RT-PCR:
-
reverse transcriptase-polymerase chain reaction.
References
Xie S, Wang Z, Okano M, Nogami M, Li Y, He WW, Okumura K, Li E: Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene. 1999, 236: 87-95. 10.1016/S0378-1119(99)00252-8.
Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP: DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000, 25: 338-342. 10.1038/77124.
Rountree MR, Bachman KE, Baylin SB: DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000, 25: 269-277. 10.1038/77023.
Fuks Francois, Hurd Paul, Deplus Rachel, Kouzarides Tony: The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003, 31: 2305-2312. 10.1093/nar/gkg332.
Melki JR, Warnecke P, Vincent PC, Clark SJ: Increased DNA methyltransferase expression in leukemia. Leukemia. 1998, 12: 311-316. 10.1038/sj.leu.2400932.
Mizuno S, Chijiwa T, Okamura T, Akashi K, Fukumaki Y, Niho Y, Sasaki H: Expression of DNA methyltransferases DNMT1, 3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood. 2001, 97: 1172-179. 10.1182/blood.V97.5.1172.
Siegel D, Hussein M, Belani C, Robert F, Galanis E, Richon VM, José GV, Cesar SR, Rizvi S: Vorinostat in solid and hematologic malignancies. J Hematol Oncol. 2009, 2: 31-10.1186/1756-8722-2-31.
Struhl K: Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998, 12: 599-606. 10.1101/gad.12.5.599.
Aggerholm A, Guldberg P, Hokland M, Hokland P: Extensive intra- and interindividual heterogeneity of p15 INK4B methylation in acute myeloid leukemia. Cancer Res. 1999, 59: 436-444.
Laura JR, Christoph P: Alterations of DNA methylation in hematologic malignancies. Cancer Lett. 2002, 185: 1-12. 10.1016/S0304-3835(02)00288-4.
Wong IH, Ng MH, Huang DP, Lee CK: Aberrant p15 promoter methylation in adult and childhood acute leukemias of nearly all morphologic subtypes: potential prognostic implications. Blood. 2000, 95: 1942-1949.
Tien HF, Tang JH, Tsay W, Liu MC, Lee FY, Wang CH, Chen YC, Shen MC: Methylation of the p15(INK4B) gene in myelodysplastic syndrome: it can be detected early at diagnosis or during disease progression and is highly associated with leukaemic transformation. Br J Haematol. 2001, 112: 148-154. 10.1046/j.1365-2141.2001.02496.x.
Ma XD, Fang YQ, Beklemisheva A, Dai W, Feng JY, Ahmed T, Liu DL, Chiao JW: Phenylhexyl isothiocyanate inhibits histone deacetylases and remodels chromatins to induce growth arrest in human leukemia cells. Int J Oncol. 2006, 28: 1287-93.
Tamaro SH, Gary DS, Mark AM, Heather Y, Mallery SR: Comparison of phenethyl and 6-phenylhexyl isothiocyanate-induced toxicity in rat esophageal cell lines with and without glutathione depletion. Toxicology Letters. 2005, 155: 427-436. 10.1016/j.toxlet.2004.11.012.
Conaway CC, Jiao Ding, Toshiyuki K, Liebes L, Chung FL: Disposition and Pharmacokinetics of Phenethyl Isothiocyanate and 6-Phenylhexyl Isothiocyanate in F344 Rats. Drug Metabolism and Disposition. 1999, 27: 13-20.
Chiao JW, Chung FL, Kancherla R, Ahmed T, Mittelman A, Conway CC: Sulforaphane and its metabolite growth arrest and apoptosis in human prostate cancer cells. Int J Oncol. 2002, 20 (3): 631-636.
Lu LL, Liu DL, Ma XD, Beklomisheva A, Seiter K, Ahmed T, Chiao JW: The phenylhexyl isothiocyanate induces apoptosis and inhibits leukemia cell growth in vivo. Oncol Reports. 2006, 16: 1363-1367.
Jiang SH, Ma XD: Advances in study for mechanism of isothiocyanates as antineoplastic agent. Chin J New Drugs Clin Rem. 2008, 27 (9): 710-713.
Talalay P, Fahey JW: Phytochemicals from cruciferous plants protect against cancer by modulating carcinogen metabolism. J Nutr. 2001, 131 (Suppl): 3027-3033.
Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB: Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. PNAS. 1996, 93 (18): 9821-9826. 10.1073/pnas.93.18.9821.
Wang LG, Liu DL, Ahmed T, Chung FL, Conaway CC, Chiao JW: Targeting cell cycle machinery as a molecular mechanism of sulforaphane in prostate cancer prevention. Int J Oncol. 2004, 24: 187-192.
Dobosy JR, Selker EU: Emerging connections between DNA methylation and histone acetylation. CMLS. 2001, 58: 721-727. 10.1007/PL00000895.
Cang SD, Ma YH, Liu DL: New clinical developments in histone deacetylase inhibitors for epigenetic therapy of cancer. J Hematol Oncol. 2009, 2: 22-10.1186/1756-8722-2-22.
Tan JH, Cang SD, Ma YH, Petrillo RL, Liu DL: Novel histone deacetylase inhibitors in clinical trials as anti-cancer agents. J Hematol Oncol. 2010, 3: 5-10.1186/1756-8722-3-5.
Zhu XP, Ma YH, Liu DL: Novel agents and regimens for acute myeloid leukemia: 2009 ASH annual meeting highlights. J Hematol Oncol. 2010, 3: 17-10.1186/1756-8722-3-17.
Moshe S, Pouya P, Shafaat AR: DNA demethylation and cancer: therapeutic implications. Cancer Lett. 2004, 211: 133-143. 10.1016/j.canlet.2004.04.009.
Ogawa M, Sakashita K, Zhao XY, Hayakawa A, Kubota Takeo, Koike K: Analysis of histone modification around the CpG island region of the p15 gene in acute myeloblastic leukemia. Leuk Res. 2007, 31: 611-621. 10.1016/j.leukres.2006.09.023.
Huang YQ, Ma XD, Zheng RJ, Chiao JW, Liu DL: Experiment study of PHI on histone methylation and actylation in Molt-4 cells. Chin J Hemato. 2007, 28 (9): 612-616.
Acknowledgements
This work was partly supported by grant-in-aid from foundation of science and technology of Zhangzhou, Fujian, China (No. Z07014), from foundation of science and technology of Fujian medical university, Fujian, China (No. FZS08018), and by grant from science research foundation of ministry of Health & United Fujian Provincial Health and Education Project for Tackling the Key Research, P.R. China. (WKJ2008-2-55).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
XM is responsible for study design, writing paper and has been involved in all aspects of this study. SJ, YH, YX and RZ contributed to collecting and analyzing data. Dr Jen-Wei contributed to the study design and manuscript preparation. All authors have read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Jiang, S., Ma, X., Huang, Y. et al. Reactivating aberrantly hypermethylated p15 gene in leukemic T cells by a phenylhexyl isothiocyanate mediated inter-active mechanism on DNA and chromatin. J Hematol Oncol 3, 48 (2010). https://doi.org/10.1186/1756-8722-3-48
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1756-8722-3-48