Abstract
Background
Pig keeping is becoming increasingly common across sub-Saharan Africa. Domestic pigs from the Arusha region of northern Tanzania were screened for trypanosomes using PCR-based methods to examine the role of pigs as a reservoir of human and animal trypanosomiasis.
Methods
A total of 168 blood samples were obtained from domestic pigs opportunistically sampled across four districts in Tanzania (Babati, Mbulu, Arumeru and Dodoma) during December 2004. A suite of PCR-based methods was used to identify the species and sub-species of trypanosomes including: Internally Transcribed Sequence to identify multiple species; species specific PCR to identify T. brucei s. l. and T. godfreyi and a multiplex PCR reaction to distinguish T. b. rhodesiense from T. brucei s. l.
Results
Of the 168 domestic pigs screened for animal and human infective trypanosome DNA, 28 (16.7%) were infected with one or more species of trypanosome; these included: six pigs infected with Trypanosoma vivax (3.6%); three with Trypanosoma simiae (1.8%); two with Trypanosoma congolense (Forest) (1%) and four with Trypanosoma godfreyi (2.4%). Nineteen pigs were infected with Trypanosoma brucei s. l. (10.1%) of which eight were identified as carrying the human infective sub-species Trypanosoma brucei rhodesiense (4.8%).
Conclusion
These results show that in Tanzania domestic pigs may act as a significant reservoir for animal trypanosomiasis including the cattle pathogens T. vivax and T. congolense, the pig pathogen T. simiae, and provide a significant reservoir for T. b. rhodesiense, the causative agent of acute Rhodesian sleeping sickness.
Similar content being viewed by others
Background
Around a third of the African continent is affected by trypanosomiasis, of which it is thought around 7 million km2 would otherwise be suitable for agricultural development[1]. African animal trypanosomiasis (AAT) is considered to be the principal disease that limits profitable and productive livestock keeping in sub-Saharan Africa[1, 2]. Domestic livestock can greatly improve the livelihoods of the rural poor, providing a wealth store that can be drawn on in times of need by selling the animal or animal products[3]. Cattle are traditionally maintained as long term investments for farmers across Sub Saharan Africa, providing draught power, manure, food and transport, which benefits agricultural development and boosts the income of rural households[4]. Cattle trypanosomiasis acts as a major impediment to economic growth in tsetse infested regions, causing severe losses in production due to poor growth, weight loss, low milk yield, reduced capacity for work, infertility and abortion[5]. Over the past decade, domestic pigs have become an increasingly popular livestock strategy in East Africa[6, 7] (see Figure 1). Free range pig production is particularly attractive to subsistence farmers as it requires low input and space, pig keeping is considered profitable since there is a perception that they demand few or even zero inputs[8, 9]. Both domestic and wild pigs can become infected with various species of tsetse transmitted trypanosomes, but infrequently show symptoms or disease pathology unless infected with Trypanosoma simiae, which is highly pathogenic for domestic pigs[10].
Estimated number of pigs in the East African countries of Burundi, Ethiopia, Kenya, Madagascar, Malawi, Mauritius, Mozambique, Reunion, Rwanda, Seychelles, Somalia, Uganda, Tanzania, Zambia, and Zimbabwe, 1961 – 2011, according to FAOSTAT online database []. http://faostat.fao.org
Human African trypanosomiasis (HAT) is the collective term for two distinct and fatal parasitic diseases, caused by two morphologically indistinguishable subspecies of Trypanosoma brucei s. l.; Trypanosoma brucei gambiense, the causative agent of chronic HAT, and T. b. rhodesiense, the causative organism of acute HAT[4, 11]. HAT causes widespread mortality and morbidity across the tsetse belt of sub-Saharan Africa, with an estimated 1.6 million disability-adjusted life years (DALYs) lost per year[12]. In the 1990s and early 2000s up to 500,000 people were considered infected with HAT with the disease being responsible for some 50,000 deaths per year[13]. In 2009 the number of HAT cases reported fell below 10,000 for the first time in 50 years, largely due to a concerted effort to address the chronic form of the disease (T. b. gambiense) led by the World Health Organization (WHO), involving many governments and non-governmental organizations[14].
Significant progress has been made reducing the burden of chronic HAT in rural Africa, which is largely managed through active human screening and case management. However, control of zoonotic T. b. rhodesiense HAT remains problematic in the absence of active screening in humans and in the animal reservoir, both of which contribute to under-reporting of the disease in humans and in animals. The long term persistence of both human infective forms in non-human reservoirs further complicates the task of assessing and reducing HAT burden and implementing control measures. These non-human reservoirs of human infective parasites serve to sustain parasite transmission in particular disease foci and maintain HAT even when active screening and treatment of human cases is in place. Persistent infection in cattle outlasts any short-term vector control strategies put in place to deal with acute HAT outbreaks[4].
HAT remains a significant public health problem in sleeping sickness foci in Tanzania. Historical records report both chronic and acute HAT cases around the shores of Lake Victoria[15–17]. Acute T. b. rhodesiense HAT is considered to have emerged in the early 1920s in Tanzania[17], although analysis of patient records in the early 1900s suggest that T. b. rhodesiense was circulating on the shores of Lake Victoria during the ‘great epidemic’[15]. Since these early large-scale epidemics the number of reported new cases of HAT in Tanzania has rarely risen above 500 per year and fell to 100 reported cases in 2009[14, 18]. Neither of these figures take into account the extensive levels of under-reporting of sleeping sickness in Tanzania[19] nor do they consider the burden on affected patients, both financially and in terms of DALYs lost[19] and there is concern that T. b. rhodesiense HAT will re-emerge as a public health problem as control measures and surveillance are relaxed[18, 20]. Public health concerns have been recently heightened by reports of drug resistant T. b. rhodesiense isolated from patients in north-western Tanzania[21] and spillover to domestic livestock and humans from wildlife reservoirs of human parasite infection in the National Parks[18]. In the district of Urambo, 98% of reported HAT cases were passively detected, and three quarters of cases were in stage two, suggesting if surveillance was carried out more cases may be detected[19].
Domestic and wild animals have been shown to harbor both T. b. gambiense and T. b. rhodesiense, but their significance in disease epidemiology remains largely un-quantified. Despite reports of T. b. gambiense in domestic (pigs, goats, sheep[22, 23]) and wild animals[24], humans are considered the primary reservoir of infection for T. b. gambiense HAT[25], responsible for disease persistence since human hosts can maintain an infection for prolonged periods of time[26].
The increasing application of molecular techniques to identify Trypanozoon isolates to the sub-species and genotype level now permits critical examination of trypanosome infection in animals and to examine the potential of various host species as reservoirs of the parasites that cause HAT infection. That the same genotype of T. b. gambiense has been isolated from humans, pigs and dogs in a HAT focus in the Democratic Republic of Congo implies inter-species transmission in some circumstances[27]. In Cameroon, T. b. gambiense was identified in pigs and small ruminants in four active HAT foci in Cameroon[22], the identification of the same T. b. gambiense genotype in man and pigs suggesting circulation between human and animals[28, 29] and supporting earlier work based on comparison of trypanosome isoenzyme profiles[30]. Tsetse blood-meal analysis also identified T. b. gambiense in the midgut of flies that had fed on human and porcine hosts[31].
Mathematical modeling based on data from sleeping sickness foci has implied that T. b. gambiense HAT could not be maintained in the focus of Bipindi without the existence of an animal reservoir[32]. However, the outcomes from these models are critically dependent on the input of length of time human and animal reservoir hosts remain infected and that parasites can be picked up by a tsetse fly. Calculating the period between infection and a patient becoming symptomatic is extremely difficult for T. b. gambiense. Domestic pigs have been shown to recover from T. b. gambiense infection, becoming trypanosome free within a year[33, 34].
For T. b. rhodesiense, an animal reservoir of infection has long been recognized as epidemiologically significant since humans become infected and die quickly and long-term infection in animal hosts maintains transmission[4]. T. b. rhodesiense has been identified in a number of epidemiological surveys that have applied PCR based methods in domestic livestock in East Africa: in cattle in Uganda[35, 36], Kenya[37] Tanzania[18]; from numerous wildlife species, in Luangwa valley in Zambia[38, 39] and in the Serengeti in Tanzania[39, 40]. Domestic cattle are long-term investments and maintain trypanosome infections for prolonged periods of time, acting as long term reservoirs of human infective parasites. Restocking of Zebu cattle in post conflict regions of Uganda has been implicated in the spread of T. b. rhodesiense HAT in Uganda, cattle restocking progressively moving the T. b. rhodesiense sleeping sickness focus northwards towards that of T. b. gambiense[41–43].
While restocking and general cattle movements have contributed to the spread of zoonotic HAT in Uganda, the role of pigs in short term transmission of T. b. rhodesiense in smallholder settings in East Africa remains less clear. Pigs are maintained for shorter periods of time but are highly susceptible to infection being frequently exposed to tsetse when free roaming[37]. Trypanosoma brucei s. l. was observed in domestic pigs in Uganda in 1984[44] but identification of human infective T. b. rhodesiense was not confirmed until pigs were sampled from three endemic foci in south-east Uganda using the blood incubation infectivity testing (BIIT)[44]. On the southern shores of lake Kyoga, Uganda, T. b. rhodesiense infection in pigs is high (1.2 - 14.6%) suggesting that pigs, although short-lived for economic reasons may play a transient role in the epidemiology of HAT[45]. In Busia, Kenya, on the eastern limits of the Ugandan T. b. rhodesiense HAT focus, pigs were shown to be at greater risk of T. b. rhodesiense infection than either cattle or small ruminants[37].
The aim of this study was to determine the role of pigs as a reservoir of infection for those trypanosomes pathogenic to other livestock (T. vivax and T. congolense in cattle) and for human pathogenic trypanosomes in the Arusha region of northern Tanzania, using PCR based methods for the detection of parasite DNA.
Methods
Samples
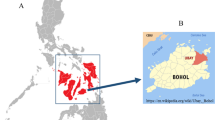
In total 168 domestic pigs were opportunistically sampled across 4 districts in Tanzania during December 2004. These comprised: nineteen pigs from Babati; seventy from Mbulu; thirty-seven from Arumeru and forty-two from Dodoma (see Figure 2). Blood samples were drawn from the jugular vein and were applied directly to Whatman FTA cards following the manufacturer’s guidelines, and air-dried at room temperature for at least 24 hrs prior to being packaged for long-term storage.
DNA preparation
Five 3 mm discs were taken from each sample and prepared for PCR following the methods previously described[45]. In brief, FTA card discs were washed twice in FTA purification reagent and twice in TE buffer, air dried before being heated at 90°C in the presence of 5% chelex solution[46]. Five microliters of FTA card eluate was used to seed each individual PCR reaction.
PCR
The following reactions were undertaken to capture the infections present in the pig samples with all reactions using primers and reaction conditions as previously published[47–50]. Internally Transcribed Sequence (ITS-PCR) were used to identify all species that were present in the sample[47]; individual species specific PCR reactions were applied to identify T. brucei s. l.[50] and T. godfreyi[49], and a multiplex PCR reaction was used to discriminate those human infective T. b. rhodesiense from T. b. brucei[51] identified from the T. brucei s. l. PCR reaction. PCR products were visualized on 1.5% agarose gels run at 100 volts for a minimum of 30 minutes using a 100 bp ladder (supperladder-mid, ABgene, UK) to size the amplified products.
Statistical analysis
Using graphpadinstat software[52], the 95% confidence interval for each percentage prevalence was computed based on the exact binomial distribution, and displayed in the relevant column in Table 1. Using the same software package the difference between prevalence of the different species and locations were compared using Fisher’s Exact Test; the differences were assigned statistical significance at p values less than 0.05. Fisher’s Exact test was used in the place of the chi squared test due to the small frequencies in some of the categories tested.
Ethical approval
The study was carried out with the full approval of pig keepers and sampling was undertaken with approval of Tanzania National Parks (TANAPA).
Results
Of the 168 domestic pig blood samples that were analyzed using the four different PCR reactions[47–51], twenty eight (16.7%) were positive for the presence of one or more trypanosome species (Table 1). Seventeen samples (10.1%) tested positive for T. brucei s. l.; the point prevalence of T. brucei s. l. being significantly lower in Mbulu than in Arumeru or Dodoma with P values of 0.0362 and 0.0387 respectively (Fishers Exact Test).
Eight (47%) of the 17 T. brucei s. l. positive pigs were identified as harboring human infective T. b. rhodesiense giving an overall prevalence of T. b. rhodesiense in pigs of 4.8%. Half of the T. b. rhodesiense infected animals identified came from the district of Arumeru, where four of 37 animals sampled were infected with T. b. rhodesiense, giving a point prevalence of 10.8%.
Application of ITS-PCR detected six cases of T. vivax (3.6%), three cases of T. simiae (1.8%) and two cases of T. congolense Forest type (1.2%) in the 168 pigs sampled; T. simiae, pathogenic to pigs, was only found in Dodoma. For T. vivax and T. congolense (Forest), the principal pathogens of cattle, there were no significant differences between districts.
Trypanosoma godfreyi was detected in 4 of the 168 domestic pig samples, generating bands at the expected size of 149 bp; again there were no significant differences between the four districts.
Mixed infections were observed in samples from 2 pigs; one pig being positive for T. vivax and T. congolense (Forest), and one pig positive for T. brucei and T. simiae. These pigs were sampled in the districts of Babati and Dodoma respectively.
Discussion
From this study it is clear that domestic pigs are making a contribution to both AAT and HAT burdens of Tanzanian livestock. Overall the prevalence of trypanosomiasis within the animals screened was 16.7%. Pigs in Arusha are acting as a significant reservoir of infection for those trypanosomes pathogenic to other livestock, T. vivax and T. congolense in cattle, sheep and goats and for the human infective parasite responsible for acute HAT, T. b. rhodesiense.
Overall, in this study ten percent of the pigs sampled were infected with T. brucei s. l. while the prevalence of T. b. rhodesiense infection in domestic pigs in this survey was high at 4.8%. Pigs sampled in Arumeru, district which lies within a sleeping sickness focus[52], had a very high prevalence of T. b. rhodesiense (10.8%).
The unusually high proportion of human infective T. b. rhodesiense circulating in domestic pigs, compared with non-human infective T. b. brucei, at almost 50% is of concern. Epidemiological theory predicts that where the non-human infective T. b. brucei and human infective T. b. rhodesiense sub-species coexist, the prevalence in non-humans of T. b. rhodesiense should exceed that of T. b. brucei[53]. However, numerous field observations using a range of methods to designate human serum resistant T. b. rhodesiense suggest the opposite to be true and show that T. b. brucei normally predominates in all domestic livestock[18, 37, 53, 54]. The overall ratio of T. b. rhodesiense to T. b. brucei in domestic livestock species was observed to be 0.35 in all studies of domestic livestock[53], the ratio that was observed in this study in Mbulu district. The very high ratios of human infective T. b. rhodesiense to non-infective T. b. brucei in pig samples in Dodoma and Arumeru, 0.428 and 0.57 respectively, mirror previous high ratios observed using the blood incubation infectivity test (BIIT) to determine human infectivity, in samples from pigs collected from a sleeping sickness endemic district of Uganda[44]. Elevated ratios of T. b. rhodesiense respective to T. b. brucei observed in domestic livestock species could be used as a One Health indicator to trigger interventions in livestock to reduce the risk to humans.
A number of studies across East Africa have implicated porcine species in the epidemiology of T. b. rhodesiense sleeping sickness; for example, a high prevalence of T. b. rhodesiense (9.5%) was identified in warthogs in the neighboring Serengeti district[18] where porcine and bovine species are the preferred hosts for local tsetse populations (Glossina pallidipes, Glossina brevipalpis and Glossina swynnertoni)[55, 56]. T. b. rhodesiense has been isolated from domestic pigs in western Kenya, on the border of the Ugandan sleeping sickness focus, where G. pallidipes are also present[57]. While data from experimental infections in pigs are available for T. b. gambiense and T. b. brucei from West Africa[33, 34], there are gaps regarding T. brucei s. l. pathology and pathogenicity in porcine hosts in East Africa including: the length of time that pigs are able to sustain infections with T. b. brucei or T. b. rhodesiense; the profile of these infections, in terms of parasitaemia, morbidity and mortality in porcine hosts and the longevity of these hosts within the farming system. Pigs are considered to be relatively short lived in small holder farming systems in East Africa[58] but may for short periods of time, contribute a significant pool of infection.
In Tanzania AAT has a large impact on animal health, second only to tick-borne East Coast Fever[59]. Cattle make up the majority of livestock kept in Tanzania and the main pathogenic species to affect cattle are Trypanosoma vivax and Trypanosoma congolense. While over 50% of the infections identified in pigs in this survey were Trypanozoon, and the prevalence of T. brucei s. l. was found to be significantly higher than that of any the other species detected, accounting for almost half of the parasites detected (17/38), T. vivax (3.6%) and T. congolense (1.2%) were also identified in pigs, which is suggestive of low rates of transmission between cattle and domestic pigs. For T. vivax and T. congolense (Forest) there were no significant differences in distribution between districts, unlike T. b. brucei and T. b. rhodesiense. T. simiae, that infects pigs, was only found in Dodoma. Sampling of pigs at the village level can be problematic for pig, vet and owner, therefore sampling of pigs at point of slaughter could provide a more straightforward strategy for assessment of HAT risk in Tanzania generally, and enable monitoring of HAT risk at district level.
Attempts in west Africa to control trypanosomiasis in pigs have shown insecticide treated nets to be an affordable and sustainable method for controlling trypanosomiasis in pigsties[60]. Pigs in east Africa are normally free ranging but a move towards simple interventions, such as housing pigs in areas where there is a risk of sleeping sickness could reduce the risk, both for trypanosomiasis, by reducing pig tsetse contact and for other zoonotic diseases (e.g. cysticercosis) by preventing contact of pigs with feces. For example, in Mbulu and Dodoma districts where T. b. rhodesiense was identified in pigs, Taenia solium cysts have also been observed in a significant number of pig carcasses[58]. Application of insecticide treatments as have been applied to prevent HAT and AAT transmission in cattle in Uganda may also offer opportunities for prevention of transmission of HAT and AAT to and from porcine hosts[61] and could be integrated with methods for control of other endemic zoonotic diseases. Approaches that simultaneously control several zoonotic diseases will amplify the impacts on animal health and pig production and human public health.
Conclusions
The prevalence of T. brucei s. l. in pigs was found to be significantly higher than that of any of the other trypanosome species, accounting for almost half of the parasites detected. That 50% of the circulating T. b. brucei s. l. identified in pigs were human infective is of public health concern, in particular the very high prevalence of T. b. rhodesiense found in Arumeru district (10.8%), which is within a sleeping sickness focus. The combination of risk factors: tsetse flies that preferentially feed on porcine hosts; pigs with a high prevalence of human infective T. b. rhodesiense; a domestic species with a high relative prevalence of T. b. rhodesiense to T. b. brucei all highlight the importance of monitoring infections in pigs in these districts. Now that PCR based methods can rapidly identify human infective parasites in animal populations, an observed high relative prevalence of T. b. rhodesiense to T. b. brucei in domestic livestock species could be applied as an indicator to trigger interventions in livestock to reduce risk to the human population.
References
Murray M, Gray AR: The current situation on animal trypanosomiasis in Africa. Prev Vet Med. 1984, 2: 23-30. 10.1016/0167-5877(84)90045-X.
Machila N, Wanyangu SW, McDermott J, Welburn SC, Maudlin I, Eisler MC: Cattle owners’ perceptions of African bovine trypanosomiasis and its control in Busia and Kwale districts of Kenya. Acta Trop. 2003, 86: 25-34. 10.1016/S0001-706X(02)00288-7.
Ellis F, Bahiigwa G: Livelihoods and rural poverty reduction in Uganda. World Dev. 2003, 31: 997-1013. 10.1016/S0305-750X(03)00043-3.
Welburn S, Coleman PG, Maudlin I, Fèvre E, Odiit M, Eisler MC: Crisis, what crisis? Control of Rhodesian sleeping sickness. Trends Parasitol. 2006, 22: 123-128. 10.1016/j.pt.2006.01.011.
Finelle P: African animal trypanosomiasis. 1983, Rome: FAO, 1-45.
FAOSTAT Online Database: [http://faostat3.fao.org/faostat-gateway/go/to/download/Q/QA/E],
Phiri IK, Ngowi H, Afonso S, Matenga E, Boa M, Mukaratirwa S, Githigia S, Saimo M, Sikasunge C, Maingi N: The emergence of Taenia solium cysticercosis in eastern and southern Africa as a serious agricultural problem and public health risk. Acta Trop. 2003, 87: 13-23. 10.1016/S0001-706X(03)00051-2.
Kagira JM, Kanyari PWN, Githigia SM, Maingi N, Ng’ang’a JC, Gachohi JM: Risk factors associated with occurrence of nematodes in free range pigs in Busia district, Kenya. Trop Anim Health Prod. 2012, 44: 657-664. 10.1007/s11250-011-9951-9.
Kagira JM, Kanyari PWN, Maingi N, Githigia SM, Ng’ang’a JC, Karuga JW: Characteristics of the smallholder free-range pig production system in western Kenya. Trop Anim Health Prod. 2010, 42: 865-873. 10.1007/s11250-009-9500-y.
Ilemobade AA, Balogun TF: Pig trypanosomiasis: effects of infection on feed intake, liveweight gain and carcass traits. Trop Anim Health Prod. 1981, 13: 128-136. 10.1007/BF02237909.
Welburn SC, Maudlin I: Priorities for the elimination of sleeping sickness. Adv Parasitol. 2012, 79: 299-337.
Fèvre EM, Wissmann B, Welburn SC, Lutumba P: The burden of human African trypanosomiasis. PLoS NTDs. 2008, 2: e333-
Welburn SC, Odiit M: Recent developments in human African trypanosomiasis. Curr Opin Infect Dis. 2002, 15: 477-484. 10.1097/00001432-200210000-00004.
Simarro P, Cecchi G, Paone M, Franco J, Diarra A, Ruiz J, Fèvre E, Courtin F, Mattioli R, Jannin J: The Atlas of human African trypanosomiasis: a contribution to global mapping of neglected tropical diseases. Int Jour Heal Geo. 2010, 9: 57-10.1186/1476-072X-9-57.
Fèvre EM, Coleman P, Welburn SC, Maudlin I: Reanalyzing the 1900–1920 sleeping sickness epidemic in Uganda. Emerg Infect Dis. 2004, 10: 567-573. 10.3201/eid1004.020626.
Koerner T, de Raadt P, Maudlin I: The 1901 Uganda sleeping sickness epidemic revisited: a case of mistaken identity?. Parasitol Today. 1995, 11: 303-306. 10.1016/0169-4758(95)80046-8.
Ormerod WE: The epidemic spread of Rhodesian sleeping sickness 1908–1960. Trans R Soc Trop Med Hyg. 1961, 55: 525-538. 10.1016/0035-9203(61)90075-X.
Kaare MT, Picozzi K, Mlengeya T, Fèvre EM, Mellau LS, Mtambo MM, Cleaveland S, Welburn SC: Sleeping sickness: a re-emerging disease in the Serengeti?. Trav Med Infect Dis. 2007, 5: 117-124. 10.1016/j.tmaid.2006.01.014.
Matemba LE, Fèvre EM, Kibona SN, Picozzi K, Cleaveland S, Shaw AP, Welburn SC: Quantifying the burden of Rhodesiense sleeping sickness in Urambo district, Tanzania. PLoS NTDs. 2010, 4: e868-
Kibona SN: Rhodesiense sleeping sickness: re-emerging as a public health problem. Tanz Heal Res Bull. 2001, 3: 48-50.
Kibona SN, Matemba L, Kaboya JS, Lubega GW: Drug-resistance of Trypanosoma b. rhodesiense isolates from Tanzania. Trop Med Int Health. 2006, 11: 144-155. 10.1111/j.1365-3156.2005.01545.x.
Njiokou F, Nimpaye H, Simo G, Njitchouang GR, Asonganyi T, Cuny G, Herder S: Domestic animals as potential reservoir hosts of Trypanosoma brucei gambiense in sleeping sickness foci in Cameroon. Parasite. 2010, 17: 61-66. 10.1051/parasite/2010171061.
Njitchouang GR, Njiokou F, Djeunga HCN, Fewou PM, Asonganyi T, Cuny G, Simo G: Analysis of the domestic animal reservoir at a micro-geographical scale, the Fontem sleeping sickness focus (south-west Cameroon). J Cell Ani Biol. 2010, 4: 73-80.
Njiokou F, Laveissère C, Simo G, Nkinin S, Grèbaut P, Cuny G, Herder S: Wild fauna as a probable animal reservoir for Trypanosoma brucei gambiense in Cameroon. Infect Gen Evol. 2006, 6: 147-153. 10.1016/j.meegid.2005.04.003.
Scott D: The Epidemiology of Gambian Sleeping Sickness. The African Trypanosomiases. Edited by: Mulligan HW. 1970, London: George, Allen and Unwin LTD, 614-644.
WHO: WHO Technical Report Series. Control and surveillance of African trypanosomiasis. 1998, Geneva: World Health Organization
Schares G, Mehlitz D: Sleeping sickness in Zaire: a nested polymerase chain reaction improves the identification of Trypanosoma (Trypanozoon) brucei gambiense by specific kinetoplast DNA probes. Trop Med Int Health. 1996, 1: 59-70. 10.1046/j.1365-3156.1996.d01-11.x.
Simo G, Asonganyi T, Nkinin SW, Njiokou F, Herder S: High prevalence of Trypanosoma brucei gambiense group 1 in pigs from the Fontem sleeping sickness focus in Cameroon. Vet Parasitol. 2006, 139: 57-66. 10.1016/j.vetpar.2006.02.026.
Simo G, Njitchouang GR, Njiokou F, Cuny G, Asonganyi T: Genetic characterization of Trypanosoma brucei circulating in domestic animals of the Fontem sleeping sickness of Cameroon. Microb Infect. 2012, 14: 651-658. 10.1016/j.micinf.2012.02.003.
Nkinin SW, Njiokou F, Penchenier L, Grabaut P, Simo G, Herder S: Characterization of Trypanosoma brucei s. l. subspecies by isoenzymes in domestic pigs from the Fontem sleeping sickness focus of Cameroon. Acta Trop. 2002, 81: 225-232. 10.1016/S0001-706X(01)00213-3.
Simo G, Silatsa B, Flobert N, Lutumba P, Mansinsa P, Madinga J, Manzambi E, De Deken R, Asonganyi T: Identification of different trypanosome species in the mid-guts of tsetse flies of the Malanga (Kimpese) sleeping sickness focus of the Democratic Republic of Congo. Parasit Vectors. 2012, 5: 201-10.1186/1756-3305-5-201.
Funk S, Nishiura H, Heesterbeek H, Edmunds WJ, Checchi F: Identifying the role of animal reservoirs in maintaining Gambiense human African trypanosomiasis. PLoS Comp Biol. 2013, 9: e1002855-10.1371/journal.pcbi.1002855.
Bungener W, Mehlitz D: Histopathological findings in mini-pigs infected with different strains of Trypanosoma brucei. Tropenmed Parasitol. 1984, 35: 109-114.
Penchenier L, Alhadji D, Bahèbèguè S, Simo G, Laveissière C, Cuny G: Spontaneous cure of domestic pigs experimentally infected by Trypanosoma brucei gambiense: implications for the control of sleeping sickness. Vet Parasitol. 2005, 133: 7-11. 10.1016/j.vetpar.2005.04.034.
Hide G, Welburn SC, Tait A, Maudlin I: Epidemiological relationships of Trypanosoma brucei stocks from South East Uganda: evidence for different population structures in human infective and non-human infective isolates. Parasitology. 1994, 109: 95-111. 10.1017/S0031182000077805.
Welburn SC, Fèvre E, Coleman P, Odiit M, Maudlin I: Sleeping sickness: a tale of two diseases. Trends Parasitol. 2001, 17: 19-24. 10.1016/S1471-4922(00)01839-0.
von Wissmann B, Machila N, Picozzi K, Fèvre EM, Bronsvoort BM d C, Handel IG, Welburn SC: Factors associated with acquisition of human infective and animal infective trypanosome infections in domestic livestock in western Kenya. PLoS NTDs. 2011, 5: e941-
Anderson NE, Mubanga J, Fèvre EM, Picozzi K, Eisler MC, Thomas R, Welburn SC: Characterisation of the wildlife reservoir community for human and animal trypanosomiasis in the Luangwa valley, Zambia. PLoS NTDs. 2011, 5: e1211-
Auty HK, Picozzi K, Malele I, Torr SJ, Cleaveland S, Welburn S: Using molecular data for epidemiological inference: assessing the prevalence of Trypanosoma brucei rhodesiense in Tsetse in Serengeti, Tanzania. PLoS NTDs. 2012, 6: e1501-
Welburn S, Picozzi K, Coleman P, Packer C: Pattern in age-seroprevalence consistent with acquired immunity against Trypanosoma brucei in Serengeti lions. PLoS NTDs. 2008, 2: e347-
Fèvre EM, Coleman P, Odiit M, Magona JW, Welburn SC, Woolhouse MEJ: The origins of a new Trypanosoma brucei rhodesiense sleeping sickness outbreak in eastern Uganda. Lancet. 2001, 358: 625-628. 10.1016/S0140-6736(01)05778-6.
Picozzi K, Févre E, Odiit M, Carrington M, Eisler MC, Maudlin I, Welburn S: Sleeping sickness in Uganda: a thin line between two fatal diseases. Br Med J. 2005, 331: 1238-1241. 10.1136/bmj.331.7527.1238.
Selby R, Bardosh K, Picozzi K, Waiswa C, Welburn SC: Cattle movements and trypanosomes: restocking efforts and the spread of Rhodesian sleeping sickness in post-conflict Uganda. Parasit Vectors. 2013, 6: 281-10.1186/1756-3305-6-281.
Okuna NM, Mayende JSP, Guloba A: Trypanosoma brucei infection in domestic pigs in a sleeping sickness epidemic area of Uganda. Acta Trop. 1986, 43: 183-184.
Waiswa C, Olaho-Mukani W, Katunguka-Rwakishaya E: Domestic animals as reservoirs for sleeping sickness in three endemic foci in south-eastern Uganda. Ann Trop Med Parasitol. 2003, 97: 149-155.
Ahmed H, MacLeod E, Hide G, Welburn S, Picozzi K: The best practice for preparation of samples from FTA(R) cards for diagnosis of blood borne infections using African trypanosomes as a model system. Parasit Vectors. 2011, 4: 68-10.1186/1756-3305-4-68.
Cox A, Tilley A, McOdimba F, Fyfe J, Eisler M, Hide G, Welburn S: A PCR based assay for detection and differentiation of African trypanosome species in blood. Exp Parasitol. 2005, 111: 24-29. 10.1016/j.exppara.2005.03.014.
Cox A, Tosas O, Tilley A, Picozzi K, Coleman P, Hide G, Welburn S: Constraints to estimating the prevalence of trypanosome infections in east African zebu cattle. Parasit Vectors. 2010, 3: 82-10.1186/1756-3305-3-82.
McNamara JJ, Mohammad G, Gibson WC: Trypanosoma (Nannomonas) godfreyi sp. nov. from tsetse flies in the Gambia: biological and biochemical characterization. Parasitology. 1994, 109: 497-509. 10.1017/S0031182000080756.
Moser DR, Cook GA, Ochs DE, Bailey CP, McKane MR, Donelson JE: Detection of Trypanosoma congolense and Trypanosoma brucei subspecies by DNA amplification using the polymerase chain reaction. Parasitology. 1989, 99: 57-66. 10.1017/S0031182000061023.
Picozzi K, Carrington M, Welburn SC: A multiplex PCR that discriminates between Trypanosoma brucei brucei and zoonotic T. b. rhodesiense. Exp Parasitol. 2008, 118: 41-46. 10.1016/j.exppara.2007.05.014.
Graphpad: QuickCalcs. [http://www.graphpad.com/quickcalcs/ConfInterval1.cfm],
Coleman PG, Welburn SC: Are fitness costs associated with resistance to human serum in Trypanosoma brucei rhodesiense?. Trends Parasitol. 2004, 20: 311-315. 10.1016/j.pt.2004.04.015.
Thumbi SM, Jung’a JO, Mosi RO, McOdimba FA: Spatial distribution of African animal Trypanosomiasis in Suba and Tesco districts in western Kenya. BMC Res Note. 2010, 3: 1-6. 10.1186/1756-0500-3-1.
Glasgow JP, Isherwood F, Lee-Jones F, Weitz B: Factors influencing the stable food of tsetse flies. J Anim Ecol. 1958, 27: 59-69. 10.2307/2174.
Weitz B, Glasgow JP: The natural hosts of some species of Glossina in east Africa. Trans R Soc Trop Med Hyg. 1956, 50: 593-612. 10.1016/0035-9203(56)90065-7.
Ng’ayo M, Njiru Z, Kenya E, Muluvi G, Osir E, Masiga D: Detection of trypanosomes in small ruminants and pigs in western Kenya: important reservoirs in the epidemiology of sleeping sickness?. Kinetoplastid Biol Dis. 2005, 4: 5-10.1186/1475-9292-4-5.
Wilson RT, Swai E: A review of pig pathology in Tanzania. Trop Anim Health Prod. 2013, In press: doi:10.1007/s11250-11013-10426-z
Mugittu KN, Silayo RS, Majiwa PAO, Kimbita EK, Mutayoba BM, Maselle R: Application of PCR and DNA probes in the characterisation of trypanosomes in the blood of cattle in farms in Morogoro, Tanzania. Vet Parasitol. 2001, 94: 177-189. 10.1016/S0304-4017(00)00365-4.
Bauer B, Holzgrefe B, Mahama CI, Baumann MPP, Mehlitz D, Clausen PH: Managing tsetse transmitted trypanosomosis by insecticide treated nets - an affordable and sustainable method for resource poor pig farmers in Ghana. PLoS NTDs. 2011, 5: e1343-
Bardosh K, Waiswa C, Welburn S: Conflict of interest: use of pyrethroids and amidines against tsetse and ticks in zoonotic sleeping sickness endemic areas of Uganda. Parasit Vectors. 2013, 6: 204-10.1186/1756-3305-6-204.
Acknowledgements
This study was supported by the European Union’s Seventh Framework Program (FP7/2007-2013) under grant agreement nº 221948 Integrated Control of Neglected Zoonoses (ICONZ) (KP, SCW); DfID, Research into Use Programme (KP, LH, SCW) and by a European Science Foundation Senior Investigator(s) award Investigating Networks of Zoonosis Innovation INZI (SCW). We would like to thank all of the many research participants that were involved in this study, the numerous government officials and others who contributed.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conceived and designed the experiment: LH, KP, MTK, SCW. Performed the experiment: LH, MTK. Analyzed the data: LH, KP. Contributed reagents/ materials/analysis tools: KP, SCW. Wrote the paper: LH, KP, SCW. All authors read and approved the final version of the MS with the exception of MTK (deceased).
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Hamill, L.C., Kaare, M.T., Welburn, S.C. et al. Domestic pigs as potential reservoirs of human and animal trypanosomiasis in Northern Tanzania. Parasites Vectors 6, 322 (2013). https://doi.org/10.1186/1756-3305-6-322
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1756-3305-6-322