Abstract
Background
B-cell precursor acute lymphoblastic leukemia (BCP-ALL) is characterized by recurrent genetic alterations including chromosomal translocations. The transcription factor PAX5, which is pivotal for B-cell commitment and maintenance, is affected by rearrangements, which lead to the expression of in-frame fusion genes in about 2.5% of the cases.
Results
Using conventional cytogenetics, fluorescence in situ hybridization (FISH), and molecular methods, an additional case with a der(9)t(7;9)(q11.23;p13) resulting in the expression of a PAX5-ELN fusion gene was identified. Furthermore, a general review of leukemia harboring a t(7;9)(q11.2;p13) or der(9)t(7;9)(q11.2;p13), which occurs more often in children than in adults and shows a remarkably high male preponderance, is given. These cytogenetically highly similar translocations lead to the expression of one of three different in frame PAX5-fusions, namely with AUTS2 (7q11.22), ELN (7q11.23), or POM121 (7q11.23), which constitute the only currently known cluster of PAX5 partner genes.
Conclusion
Our report underlines the recurrent involvement of PAX5 in different fusion genes resulting either from t(7;9)(q11.2;p13) or der(9)t(7;9)(q11.2;p13), which cannot be distinguished cytogenetically and whose discrimination requires molecular analysis.
Similar content being viewed by others
Background
PAX5 rearrangements, resulting in the expression of in-frame fusion genes, account for about 2.5% of pediatric B-cell precursor acute lymphoblastic leukemia (BCP-ALL) [1]. While several groups, including our own, have reported the incidence and diversity of PAX5 fusion genes [1–7], their occurrence in leukemia harboring a t(7;9)(q11.2;p13) or der(9)t(7;9)(q11.2;p13) has not yet been investigated in detail. Herein, we describe an additional case with a PAX5-ELN fusion and summarize the demographic and genetic data of all cases with t(7;9)(q11.2;p13)/der(9)t(7;9)(q11.2;p13) leukemia reported to date.
Case presentation
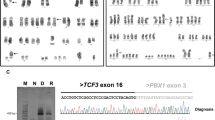
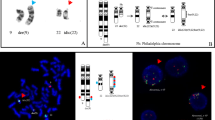
We have identified an additional case of pediatric BCP-ALL with an infrequent der(9)t(7;9)(q11.23;p13) resulting in the expression of an in-frame PAX5-ELN fusion gene (Table 1). Cytogenetic analysis of the bone marrow of a 19.4-year-old adolescent revealed - together with many secondary aberrations - a der(9)t(7;9)(q11.2;p13) (Figure 1A) and subsequent FISH analysis using PAX5-flanking BAC clones showed a deletion of the 3′-end-specific probe, which is suggestive of a PAX5 gene rearrangement (Figure 1B). Further FISH analysis, using PAX5- and ELN-specific clones, identified ELN as the fusion partner (Figure 1C), which was verified on the molecular level by RT-PCR (Figure 1D). Sequencing of the amplification product showed that exon 7 of PAX5 was fused to exon 5 of ELN (Figure 1E). In all PAX5-ELN cases, except for one in which PAX5 exon 5 was fused to ELN sequences, the breakpoints in PAX5 occurred in intron 7 [2, 4, 8]. Also, the breakpoints in ELN are heterogeneous and PAX5 is fused to either exon 2 or exon 5 of ELN (Table 1) [2, 4, 8]. Consequently, the consensus PAX5-ELN fusion protein consists of the DNA-binding paired domain, the octapeptide, and the nuclear localization signal of PAX5, which are fused to almost the entire ELN protein without the signal peptide (Figure 2).
Cytogenetic and molecular genetic analysis of a PAX5-ELN positive case. (A) Karyogram; red arrow indicates the derivative chromosome der(9)t(7;9)(q11.23;p13) (refined karyotype using molecular methods). (B) FISH using PAX5-specific BAC clones showing a 3′-end deletion: 5′-end-specific clone (red signals); 3′-end-specific clone (green signals); black and red arrows indicate the normal and derivative chromosome, respectively. (C) FISH using PAX5- and ELN-specific BAC clones showing a co-localization: PAX5 5′-end-specific clone (red signals); ELN 3′-end-specific clone (green signals); arrows indicate the normal chromosomes 9 and 12 (white) and the derivative chromosome (black). (D) RT-PCR using primers located in PAX5 exon 2–3 and ELN exon 6 resulting in amplification of PAX5-ELN fusion transcripts. M, molecular weight marker DNA-mix ladder (Peqlab); lane 1, patient No. 5; lane 2, normal control. (E) Sequence chromatogram of the PAX5-ELN fusion junction showing the fusion between exon 7 of PAX5 and exon 5 of ELN.
Schematic representation of the structure of PAX5 and the putative consensus chimeric proteins. PD, paired domain; 8, octapeptide; HD, partial homeodomain; TA, transactivation domain; I, inhibitory domain; P, proline-rich regions; H, histidine-rich regions; KA, alanine-rich cross-linking domains; KP, proline-rich cross-linking domains; HY, hydrophobic domains; 6, VGVAPG hexapeptide domain; C, C-terminal domain; i, insertion; N, POM121 5′-untranslated region; FG, FG-repeats; arrows and filled lollipops indicate nuclear localization signals and fusion breakpoints, respectively.
Results and discussion
So far, sixteen in-frame PAX5-fusions have been described and the fusion partners comprise a heterogeneous group of genes encoding proteins, which play distinct roles in signaling, transcription, chromatin remodeling, and cell structuring [1–7, 9, 10]. Three of the sixteen currently known PAX5 fusion partners, namely, AUTS2, ELN, and POM121, are located at the pericentromeric region of 7q and encompass roughly 3.3 Mb of genomic DNA, forming the only currently known cluster of PAX5 partner genes. Therefore, t(7;9)(q11.2;p13) translocations may give rise to three different recurrent fusion genes, i.e. PAX5-AUTS2, PAX5-POM121, and PAX5-ELN (Figure 2), which are not distinguishable at the cytogenetic level. In addition, karyotyping of five of the cases showed an unbalanced der(9)t(7;9)(q11.2;p13) with loss of the reciprocal derivative chromosome (Table 1; cases 5–7 and 11–12). One of the cases was identified by SNP array (case 4), only detecting unbalanced chromosome alterations; furthermore, most cases showed a deletion of the PAX5 3′-end by FISH, further supporting the notion that the PAX5- partner fusions, and not the reciprocal ones contribute to leukemogenesis.
t(7;9)(q11.2;p13)/der(9)t(7;9)(q11.2;p13) positive leukemia is a rare disease and only 9 cases have been collected in the Mitelman database of the cancer genome anatomy project ([11] accessed November 2013) (Table 1). Three of these cases were PAX5-ELN positive [2, 4] and in addition, a case with a PAX5-ELN fusion without cytogenetic data has been reported [8]. Together, including the case described herein, five patients harboring this fusion gene have now been identified.
Other cases involving the cluster of PAX5 fusion partners include: Three patients with a PAX5-AUTS2[5, 9, 10], two with a PAX5-POM121 fusion gene [1, 4], and in two cases involvement of PAX5 has not been investigated [12, 13] (Table 1). Of note, in the PAX5-POM121 case we have previously published [1], cytogenetic analysis failed to identify a t(7;9)(q11.2;p13), but the chromosome quality was rather poor. Whole chromosome painting with probes specific for chromosomes 7 and 9 showed the presence of a der(7;9), on which the 3′-end of PAX5 was located, whereas the 5′-end of PAX5, generating the PAX5-POM121 fusion, was translocated to a derivative chromosome, which only partially consisted of chromosome 7 material (data not shown). Together, with the molecular data that showed an insertion of chromosome 12 sequences in the fusion, a more complex rearrangement with involvement of at least chromosomes 7, 9, and 12 generated the in-frame PAX5-POM121 fusion [1].
Furthermore, out of the 12 cases with t(7;9)(q11.2;p13)/der(9)t(7;9)(q11.2;p13) rearrangements only one was an adult and two were young adolescents, whereas all other patients were ≤ 4 years of age (Table 1), suggesting that this subtype of leukemia occurs more frequently in pediatric than in adult cases. Remarkably, 83% (10/12) of the t(7;9)(q11.2;p13)/der(9)t(7;9)(q11.2;p13) patients were male, and thus, the male/female ratio was 5. Although the number of so far reported cases is rather low, in acute leukemia such an extreme gender bias is exceedingly rare [14]. This finding is intriguing, but currently there is no plausible explanation why a specific subtype of leukemia is associated with one or the other gender.
Regarding the prognostic relevance of PAX5 fusion genes in general, due to their rareness no final conclusions may be drawn. However, we have recently shown that PAX5-AUTS2 leukemia may have a rather unfavorable outcome [10]. Out of the five PAX5-ELN cases, one patient (case 4) showed high-risk features and displayed a JAK1 mutation and a BCR-ABL1-like expression signature [8]. Furthermore, cases 1 and 2 both relapsed post allograft and died 16 months after initial diagnosis ([15] accessed November 2013). The PAX5-ELN positive patient presented herein is currently, eight months after initial diagnosis, in complete remission. Together, there is at least some evidence that t(7;9)(q11.2;p13)/der(9)t(7;9)(q11.2;p13) leukemia may have a rather poor prognosis. However, whether this is attributable to the specific PAX5-fusions or to coinciding mutations in, for example, tyrosine kinases, remains to be determined, and a larger cohort of patients needs to be analyzed, which, due to the low incidence of this leukemia subtype, will require an international collaborative effort.
Conclusion
In this report an additional case of PAX5-ELN positive leukemia is described, and, furthermore, an overview of the published cases of t(7;9)(q11.2;p13)/der(9)t(7;9)(q11.2;p13) leukemia is given, emphasizing the importance of molecular analysis to discriminate between cytogenetically identical translocations resulting in distinct fusion genes.
Material and methods
Cytogenetic and fluorescence in situ hybridization (FISH) analysis
Cytogenetic analysis was performed according to standard techniques. FISH analysis using PAX5- and ELN-specific probes was conducted as previously described [1]. The PAX5 rearrangement was first detected using PAX5-flanking BAC clones RP11-220I1 and RP11-12P15 (obtained from Pieter de Jong, BACPAC Resources, Children’s Hospital and Research Center Oakland, CA, USA). Verification of the PAX5-ELN fusion was performed using the PAX5 5′-end-flanking BAC clone RP11-220I1 in combination with the ELN 3′-end-specific clone RP11-349P21 (Welcome Trust Sanger Institute; http://www.sanger.ac.uk).
RNA isolation and reverse transcription-polymerase chain reaction (RT-PCR)
RNA isolation and RT-PCR for the detection of PAX5-ELN transcripts were performed according to standard procedures using primers PAX5ex2-3-F1 (5′-TCTTGGCAGGTATTAT GAGACAGGAAG-3′) and ELNex6-R2 (5′-AGCAGCGTCAGCCACTCCAC-3′) located in exons 2–3 and 6 of PAX5 and ELN, respectively. Amplification products were directly sequenced (Microsynth AG, Austria) and sequence analysis was conducted using the CLC Main Workbench 6.0 (CLC bio, Denmark).
Reference sequences and exon nomenclature
The chromosome band positions of the genes and the exon nomenclature used correspond to that of the Ensemble database and the reference sequences for AUTS2 (ENST00000342771), ELN (ENST00000252034), POM121 (ENST00000257622), and PAX5 (ENST00000358127) (Ensembl release 73 - September 2013). A summary of all mRNA fusion sequences as well as the entire transcript and protein sequences of the putative consensus chimeras PAX5-AUTS2, PAX5-ELN, and PAX5-POM121 are provided as Additional file 1.
Consent
Within the AIEOP-BFM ALL 2009 study (ClinicalTrials.gov Identifier: NCT01117441), written informed consent - which includes the compliance that surplus material not required for diagnostic purposes may be used for research purposes - is obtained from the patients, their parents or their legal guardians. This study has exclusively been performed on material obtained for diagnostic purposes and neither any additional medical intervention nor patient recruitment was necessary.
References
Nebral K, Denk D, Attarbaschi A, Konig M, Mann G, Haas OA, Strehl S: Incidence and diversity of PAX5 fusion genes in childhood acute lymphoblastic leukemia. Leukemia 2009, 23: 134–143. 10.1038/leu.2008.306
Bousquet M, Broccardo C, Quelen C, Meggetto F, Kuhlein E, Delsol G, Dastugue N, Brousset P: A novel PAX5-ELN fusion protein identified in B-cell acute lymphoblastic leukemia acts as a dominant negative on wild-type PAX5. Blood 2007, 109: 3417–3423. 10.1182/blood-2006-05-025221
Cazzaniga G, Daniotti M, Tosi S, Giudici G, Aloisi A, Pogliani E, Kearney L, Biondi A: The paired box domain gene PAX5 is fused to ETV6/TEL in an acute lymphoblastic leukemia case. Cancer Res 2001, 61: 4666–4670.
Coyaud E, Struski S, Prade N, Familiades J, Eichner R, Quelen C, Bousquet M, Mugneret F, Talmant P, Pages MP, et al.: Wide diversity of PAX5 alterations in B-ALL: a groupe francophone de cytogenetique hematologique study. Blood 2010, 115: 3089–3097. 10.1182/blood-2009-07-234229
Kawamata N, Ogawa S, Zimmermann M, Niebuhr B, Stocking C, Sanada M, Hemminki K, Yamatomo G, Nannya Y, Koehler R, et al.: Cloning of genes involved in chromosomal translocations by high-resolution single nucleotide polymorphism genomic microarray. Proc Natl Acad Sci U S A 2008, 105: 11921–11926. 10.1073/pnas.0711039105
Lee ST, Ji Y, Kim HJ, Ki CS, Jung CW, Kim JW, Kim SH: Sequential array comparative genomic hybridization analysis identifies copy number changes during blastic transformation of chronic myeloid leukemia. Leuk Res 2012, 36: 418–421. 10.1016/j.leukres.2011.12.021
Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, et al.: Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 2007, 446: 758–764. 10.1038/nature05690
Mullighan CG, Zhang J, Harvey RC, Collins-Underwood JR, Schulman BA, Phillips LA, Tasian SK, Loh ML, Su X, Liu W, et al.: JAK mutations in high-risk childhood acute lymphoblastic leukemia. PNAS Supplements 2009, 106: 9414–9418. 10.1073/pnas.0811761106
Coyaud E, Struski S, Dastugue N, Brousset P, Broccardo C, Bradtke J: PAX5-AUTS2 fusion resulting from t(7;9)(q11.2;p13.2) can now be classified as recurrent in B cell acute lymphoblastic leukemia. Leuk Res 2010, 34: e323-e325. 10.1016/j.leukres.2010.07.035
Denk D, Nebral K, Bradtke J, Pass G, Moricke A, Attarbaschi A, Strehl S: PAX5-AUTS2: a recurrent fusion gene in childhood B-cell precursor acute lymphoblastic leukemia. Leuk Res 2012, 36: e178-e181. 10.1016/j.leukres.2012.04.015
Mitelman F, Johansson B, Mertens F: Mitelman database of chromosome aberrations and gene fusions in cancer. 2013.http://cgap.nci.nih.gov/Chromosomes/Mitelman
Raimondi SC, Zhou Y, Mathew S, Shurtleff SA, Sandlund JT, Rivera GK, Behm FG, Pui CH: Reassessment of the prognostic significance of hypodiploidy in pediatric patients with acute lymphoblastic leukemia. Cancer 2003, 98: 2715–2722. 10.1002/cncr.11841
van Zutven LJ, van Drunen E, de Bont JM, Wattel MM, Den Boer ML, Pieters R, Hagemeijer A, Slater RM, Beverloo HB: CDKN2 deletions have no prognostic value in childhood precursor-B acute lymphoblastic leukaemia. Leukemia 2005, 19: 1281–1284. 10.1038/sj.leu.2403769
Cartwright RA, Gurney KA, Moorman AV: Sex ratios and the risks of haematological malignancies. Br J Haematol 2002, 118: 1071–1077. 10.1046/j.1365-2141.2002.03750.x
Bousquet M, Dastugue N, Brousset P: t(7;9)(q11;p13). 2007.http://AtlasGeneticsOncology.org/Anomalies/t0709q11p13ID1195.html
Acknowledgements
We thank Andishe Attarbaschi, Gertrud Pass, and Klaus Fortschegger for helpful discussions. This work was supported by a grant from the Austrian Science Fund (FWF P21554-B19 to S.S.) and the St. Anna Kinderkrebsforschung e.V.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interest.
Authors’ contributions
DD conducted experiments, analyzed the data, and wrote the manuscript; MK conducted FISH analysis; JB performed cytogenetic analysis; SS supervised the study and drafted the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
13039_2013_362_MOESM1_ESM.docx
Additional file 1: PAX5 fusion genes in t(7;9)(q11;p13) leukemia: A case report and review of the literature. (DOCX 45 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Denk, D., Bradtke, J., König, M. et al. PAX5 fusion genes in t(7;9)(q11.2;p13) leukemia: a case report and review of the literature. Mol Cytogenet 7, 13 (2014). https://doi.org/10.1186/1755-8166-7-13
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1755-8166-7-13