Abstract
Background
Little genomic or trancriptomic information on Ganoderma lucidum (Lingzhi) is known. This study aims to discover the transcripts involved in secondary metabolite biosynthesis and developmental regulation of G. lucidum using an expressed sequence tag (EST) library.
Methods
A cDNA library was constructed from the G. lucidum fruiting body. Its high-quality ESTs were assembled into unique sequences with contigs and singletons. The unique sequences were annotated according to sequence similarities to genes or proteins available in public databases. The detection of simple sequence repeats (SSRs) was preformed by online analysis.
Results
A total of 1,023 clones were randomly selected from the G. lucidum library and sequenced, yielding 879 high-quality ESTs. These ESTs showed similarities to a diverse range of genes. The sequences encoding squalene epoxidase (SE) and farnesyl-diphosphate synthase (FPS) were identified in this EST collection. Several candidate genes, such as hydrophobin, MOB2, profilin and PHO84 were detected for the first time in G. lucidum. Thirteen (13) potential SSR-motif microsatellite loci were also identified.
Conclusion
The present study demonstrates a successful application of EST analysis in the discovery of transcripts involved in the secondary metabolite biosynthesis and the developmental regulation of G. lucidum.
Similar content being viewed by others
Background
Ganoderma lucidum (Curtis: Fr.) P. Karst, Lingzhi in Chinese, which belongs to the Polyporaceae family, has been used in China as medicine for centuries to promote health and longevity [1, 2]. In other countries, its fruiting body is used to treat a variety of ailments, such as cancers, hypertension, diabetes, and hepatitis, apart from being a dietary supplement [2–4]. G. lucidum is an anti-tumour agent that acts via immune modulation or stimulating cytokine production [5–7]. The bioactive constituents of G. lucidum include more than 120 different triterpenes and polysaccharides, proteins and other compounds [2, 8].
Genes involved in the triterpenoids biosynthesis pathways in G. lucidum including squalene synthase (SQS), farnesyl-Diphosphate Synthase (GlFPS) and HMG-CoA reductase (Gl -HMGR) were isolated and characterized [9–11]. Joo et al. identified a laccase gene (GLLac1) from G. lucidum[12]. However, little is known about the molecular biology of its fruiting body and its secondary metabolism. Identification of expressed genes, in particular the transcript profile, of the G. lucidum fruiting body would be a key to understanding its molecular biology.
Expressed sequence tag (EST) analysis allows rapid and large-scale identification of uniquely expressed genes [13, 14]. The EST analysis was used in transcriptome analysis of Lentinula edode[15], Aspergillus niger[16], Ustilago maydis[17] and Neurosphora crassa[18]. Sequencing information from ESTs may help discover genes in the biosynthesis of secondary metabolites [19]. Loo et al. identified a gene involved in the ricinoleic acid biosynthetic pathway [20]. Recently, genes encoding enzymes involved in the biosynthesis of ginsenoside, triterpene saponin and diterpenes were identified [21–23]. EST sequencing identified simple sequence repeats (SSRs) for genetic mapping [24].
Using the EST analysis, the present study annotated functional genes involved in the biosynthesis of secondary metabolites and the developmental regulation of the fruiting body of G. lucidum. Unique sequences very similar to squalene epoxydase (SE) and farnesyl-diphosphate synthase (FPS) in this EST collection were identified. We also discussed several candidate transcripts possibly associated with the cellular development of G. lucidum, such as hydrophobin, MOB2, profilin and PHO84. Moreover, identifying SSRs in the EST data is useful in marker-assisted breeding programs.
Methods
RNA extraction and cDNA library construction
The fruiting body of G. lucidum was obtained from the co-author Jin Lan, who has long been engaged in Ganoderma research in the Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China. She authenticated the G. lucidum using the morphological identification approach and referred to the Fungi Identification Manual [25]. Fifty (50) days after growing on the basswood medium at 25-30°C in a shade shelter, approximately 0.5 g was harvested and frozen in liquid nitrogen immediately. The mRNA of thefruiting body was isolated and purified directly with a Dynabeads (R) mRNA DIRECT™ kit (Invitrogen, USA) according to the manufacturer's recommendations. The cDNA library was constructed from purified mRNA with a Creator™ SMART™ cDNA Library Construction kit (Clontech, USA). The double-stranded cDNA was directionally ligated into the Sfi I restriction site of the pDNR-lib vector (Clontech, USA) and electroporated into a DH5α Escherichia coli strain (TakaRa, Japan).
EST sequencing, assembly and annotation
A total of 1,023 randomly selected clones were cultured in liquid LB medium containing 34 mg/l chloramphenicol and incubated overnight at 220 rpm in rotation and 37°C. Plasmid DNA was prepared with an Axyprep-96 plasmid kit (Axygen, USA). The plasmid DNA was submitted for direct sequencing from the 5' end with an M13 forward primer on an ABI 3730 DNA sequencer using BigDye 3.1 sequencing chemistry (Applied Biosystems, USA).
The ABI-formatted chromatogram sequences were processed automatically with a local EST analysis pipeline. The Phred/Phrap program was applied for trace files conversion and for base calling with quality assessment [26, 27]. The vector and low-quality regions were removed from the sequence with the Cross Match typically included in the Phred/Phrap program. The short sequences (less than 100 bp) and poly A/T tails were filtered from the EST database. The high quality ESTs were assembled into contigs (clusters of assembled ESTs) and singletons (sequences found only once) by Phrap [28].
The unique sequences were searched against public databases including the SwissProt [29], NCBI non-redundant protein (Nr) [30] and non-redundant nucleotide (Nt) [31] databases using BLAST [32] algorithm, with a E-value cut-off at 10-5. The functional categories of these unique sequences were classified by a broad category, including metabolism, energy production, cell signalling, cell defence and stress response, cell structure and growth, transcription, protein synthesis, protein degradation, transport and secretion as well as unclassified and unknown function.
SSR detection
The detection of simple sequence repeats (SSRs) from the total high-quality ESTs of the fruiting body of G. lucidum was performed with the Simple Sequence Repeat Identification Tool (SSRIT) [33]. The SSRIT accepts FASTA-formatted sequence files and reports the sequence ID, SSR motif, number of repeats (di- and tri-nucleotide repeat units), repeat length and position of the SSR and the total length of the sequence in which the SSRs were found [34]. The search parameters for the maximum motif-length group were set to hexamer and those for the minimum number of repeats were set to five.
Result and discussion
General characteristics of G. lucidum fruiting body cDNA library and ESTs
A cDNA library was constructed from the fruiting body of G. lucidum for the identification of the transcripts and the expression profiles involved in its cellular development and biosynthesis of secondary metabolites. The cDNA library had a titre of 1.25×106 colony forming units per millilitre (ml). A total of 1,023 cDNA clones were randomly selected from the library for sequencing, yielding 879 (85.9%) high-quality ESTs after vector screening and short sequence (<100 bp) filtering. These ESTs were assembled into 82 contigs and 518 singletons for a total of 600 unique sequences (Table 1). The average sequence length of these unique sequences was 288 bp, ranging from 0.15 kb to 1.5 kb. Over 63.4% contigs had two sequences, followed by 22.0% having three to four sequences and 14.6% having five to 40 sequences. Approximately 31.75% of the ESTs were redundant. A total of 427 unique sequences (71.1%) displaying no similarities to any sequences in the public databases were probably new transcripts. The redundancy (31.75%) of the ESTs suggests considerable potential for new transcripts in continued sequencing of random colonies from this cDNA library. The sequenced transcripts have been deposited in the GenBank database (GO447131-GO448009).
Expressed profile of the unique sequences
The expressed profile of the unique sequences identified in the G. lucidum fruiting body is shown in Table 2. Among 600 unique sequences, 518 (86.3%) unique sequences were sequenced only once; 72 (12%) unique sequences 2-5 times; five (0.8%) unique sequences 6-10 times and five (0.8%) unique sequences 12 times or more. The most abundantly expressed unique sequences in the G. lucidum fruiting body were coded for the hypothetical proteins (48 ESTs) and the cell wall-associated hydrolase (36 ESTs) (Table 3). Moreover, the unique sequence consisting of five ESTs and with sequence similarity to hydrophobin 2 of Lentinula edodes (Xianggu) was identified for the first time in G. lucidum (Table 3). Moreover, the unique sequences matched the elongation factors and the ribosomal proteins were also expressed at high levels (Table 3).
Annotation of expressed sequence tags
The list of the annotated ESTs found in the fruiting body of G. lucidum is shown in Additional file 1. Sixty-two (62) ESTs showed sequence similarities to uncharacterized genes encoding hypothetical proteins that were omitted from the list. The unique sequences from this cDNA library were analyzed for similarities by performing BLAST searches against public databases, including SwissProt [29], Nr [30] and Nt [31]. A total of 139 (23.2%) and 67 (11.2%) unique sequences were assigned a putative identity based on significant sequence similarities to at least one sequence in the Nr and Nt databases, respectively. These annotated unique sequences provide an available resource for application and basic microbiology. Furthermore, among the 879 ESTs only three (0.3%) ESTs were identified as homologues of previously reported nucleotides from G. lucidum in the GenBank database, indicating that the vast majority of the ESTs in our dataset were unique and new. The three unique sequences showed similarities to cytochrome c oxidase subunit 2 (GO447869), glyceraldehyde-3-phosphate dehydrogenase (GO447698) and FPS (GO447502) (Additional file 1).
Functional distribution of ESTs
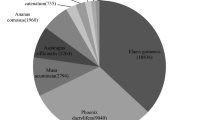
The functions of the proteins that the identified sequences encoded were classified into categories of metabolism, energy production, cell signalling, cell defence and stress response, cell structure and growth, transcription, protein synthesis, protein degradation, transport and secretion as well as unclassified and unknown functions. The functional distribution of identified sequences from the G. lucidum fruiting body cDNA library is shown in Figure 1. The unique sequences associated with metabolism (22%), protein synthesis (22.7%), unclassified and unknown function (12.7%) and energy production (11.3%) were strongly represented, whereas those with cell signalling (2.0%) and cell defence and stress response (3.3%) were not. In summary, a total of 173 unique sequences showed similarities to known genes involved in the biosynthesis of secondary metabolites and developmental regulation. While the EST sequencing scale is limited, it provides some information about the expressed transcript profile of the fruiting body of G. lucidum.
Distribution of ESTs by broad functional categories. The functional categories include: metabolism, energy production, cell signaling, cell defence and stress response, cell structure and growth, transcription, protein synthesis, protein degradation, transport and secretion, and unclassified and unknown function.
SSR detection
SSRs, also known as microsatellites, are useful genetic markers in molecular biology. A total of 13 SSR motifs were identified from the EST sequences of the fruiting body of G. lucidum (Table 4). The composition of di- and tri-nucleotide SSRs included AC (two), AT (two), CT (one), GA (one), GT (one), TA (one), TC (one), and TG (two). Only two tri-nucleotide repeats with the composition of CGA and GGT were found in the EST dataset (Table 4). Each SSR-containing unique sequence contains only one SSR motif. The lengths of eight repeats were nine bases and the other five repeats were between 11 and 15 bases. In addition, the tetra-, penta- and hexa-nucleotide repeats were not present in this EST dataset. There was a wide variation in the frequency of SSR motifs among species [35].
Candidate genes involved in the biosynthesis of triterpenoids
EST analysis is an important tool to identify secondary metabolite genes in the fruiting body of G. lucidum. Triterpenoids, the major bioactive compounds in G. lucidum, are synthesized from acetyl-CoA in the isoprenoid pathway. While genes involved in the triterpenoid biosynthetic pathway including SQS, GlFPS and Gl-HMGR were cloned from and identified in G. lucidum[9–11], other genes for the key enzymes in this pathway are to be identified.
According to the studies of the triterpene biosynthesis [10, 36], SE and FPS are rate-limiting enzymes in catalyzing triterpenoid biosynthesis in G. lucidum. The unique sequence (GO447913) with 71% identity (E-value = 1.00-12) to SE and the unique sequence (GO447502) with 98% identity to FPS (E-value = 4.00-27) involved in triterpenoid biosynthesis were presented in our EST data. SE acts as an important regulatory enzyme in the triterpenoid biosynthetic pathway. SE (EC 1.14.99.7), a monooxygenase, converts squalene into 2,3-oxidosqualene [36, 37]. The enzyme requires molecular oxygen, flavin adenine dinucleotide (FAD), either NADH or NADPH depending on the organisms [38]. Since the gene encoding SE has not been identified in G. lucidum, the information of the unique sequence (GO447913) will help identify and characterize the SE in G. lucidum. The EST for FPS (GO447502) shows sequence similarity to the GlFPS, suggesting that this EST is the partial sequence of the full-length GlFPS.
The genes encoding key enzymes involved in the triterpenoid biosynthesis, such as SQS, Gl-HMGR and others, are not present in this EST dataset, indicates a low abundance of these genes in the fruiting body of G. lucidum or incomplete sequencing of the library. The absence of ESTs associated with the polysaccharide biosynthesis, which should be abundant in G. lucidum, in this study may be due to the limited sequencing scale.
Candidate genes involved in regulation of G. lucidum development
Several transcripts present in the EST dataset encode the proteins that may be associated with the development processes of the fruiting body of G. lucidum. The unique sequence (GO447641) showed sequence similarity to the inorganic phosphate transporter PHO84 gene which controls the absorption of phosphate nutrition and regulates the development of Saccharomyces cerevisiae[39]. MOB2 is a nonessential yeast gene and plays a role in the maintenance of ploidy [40]. The unique sequences homologous to MOB2 (GO447972) and PHO84 (GO447641) may have the same functions as those in yeast. Hydrophobin is expressed specially in filamentous fungi and is important during the morphogenesis of fungi and the fruiting body development of mushrooms [41]. The unique sequence homologous to hydrophobin 2 of Lentinula edodes in this cDNA library consisted of five ESTs (GO447695, GO447166, GO447364, GO447414, GO447512), suggesting its abundance in the fruiting body of G. lucidum. Suizu et al. (2008) reported that the ESTs for hydrophobins were also most frequently identified in the cDNA library of Lentinula edodes[15]. Profilin is a universal small eukaryotic protein that binds to monomeric actin (G-actin) and is involved in diverse functions such as maintenance of cell structural integrity, cell mobility and growth factor signal transduction [42]. The sequences (GO447955, GO447282) encoding profilin were present in the G. lucidum cDNA library. The important unique sequence encoding an argonaute-like protein (GO447302) may be involved in the RNAi pathway, suggesting a potential gene knock-out by RNA interference in G. lucidum. Cloning and characterization of these candidate genes is under way.
Limitations of the study
The ESTs sequenced in this study from the fruiting body of G. lucidum were insufficient to cover all functional genes, although this EST dataset showed some characteristics of gene expression in the fruiting body of G. lucidum.
Conclusion
The present study used EST analysis and identified the transcripts in the biosynthesis of secondary metabolites and the developmental regulation of G. lucidum. For example, the candidate transcript encoding SE, the rate-limiting enzyme in the triterpenoid biosynthesis, was identified. Several genes associated with the development processes of G. lucidum, such as hydrophobin, MOB2, profilin and PHO84, were also identified.
Abbreviations
- BLAST:
-
Basic Local Alignment Search Tool
- bp:
-
base pair
- cDNA:
-
complementary DNA
- EST:
-
expressed sequence tag
- FPS:
-
farnesyl-diphosphate synthase
- HMGR:
-
HMG-CoA reductase
- NCBI:
-
National Center for Biotechnology Information
- Nr:
-
NCBI non-redundant protein
- Nt:
-
NCBI non-redundant nucleotide
- SE:
-
squalene epoxidase
- SQS:
-
squalene synthase
- SSRs:
-
simple sequence repeats
References
Wachtel-Galor S, Tomlinson B, Benzie IFF: Ganoderma lucidum ("Lingzhi"), a Chinese medicinal mushroom: biomarker responses in a controlled human supplementation study. Br J Nutr. 2004, 91: 263-269. 10.1079/BJN20041039.
Paterson MRR: Ganoderma - A therapeutic fungal biofactory. Phytochemistry. 2006, 67: 1985-2001. 10.1016/j.phytochem.2006.07.004.
Yun TK: Update from Asia. Asian studies on cancer chemoprevention. Ann NY Acad Sci. 1999, 889: 157-192. 10.1111/j.1749-6632.1999.tb08734.x.
Sliva D: Cellular and physiological effects of Ganoderma lucidum (Reishi). Mini-Rev Med Chem. 2004, 4: 873-879.
Wang SY, Hsu ML, Hsu HC, Tzeng CH, Lee SS, Shiao MS, Ho CK: The anti-tumor effect of Ganoderma lucidum is mediated by cytokines released from activated marophages and tlymphocytes. Int J Cancer. 1997, 70: 699-705. 10.1002/(SICI)1097-0215(19970317)70:6<699::AID-IJC12>3.0.CO;2-5.
Lin ZB, Zhang HN: Anti-tumor and immunoregulatory activities of Ganoderma lucidum and its possible mechanisms. Acta Pharmacol Sin. 2004, 25: 1387-1395.
Lee SS, Wei YH, Chen CF, Wang SY, Chen KY: Anti-tumor effects of Ganoderma lucidum. J Chin Med. 1995, 6: 1-12.
Kim HW, Kim BK: Biomedicinal triterpenoids of Ganoderma lucidum (Curt.:Fr.) P. Karst. (aphyllophoromycetideae). Int J Med Mushrooms. 1999, 1: 121-138.
Zhao MW, Liang WQ, Zhang DB, Wang N, Wang CG, Pan YJ: Cloning and characterization of squalene synthase (SQS) gene from Ganoderma lucidum. J Microbiol Biotechnol. 2007, 17: 1106-1112.
Ding YX, Ou-Yang X, Shang CH, Ren A, Shi L, Li YX, Zhao MW: Molecular cloning, characterization, and differential expression of a farnesyl-diphosphate synthase gene from the Basidiomycetous fungus Ganoderma lucidum. Biosci Biotechnol Biochem. 2008, 72: 1571-1579. 10.1271/bbb.80067.
Shang CH, Zhu F, Li N, Ou-Yang X, Shi L, Zhao MW, Li YX: Cloning and characterization of a gene encoding HMGR-CoA reductase from Ganoderma lucidum and its functional identification in yeast. Biosci Biotechnol Biochem. 2008, 72: 1333-1339. 10.1271/bbb.80011.
Joo SS, Ryu IW, Park JK, Yoo YM, Lee DH, Hwang KW, Choi HT, Lim CJ, Lee DI, Kim KH: Molecular cloning and expression of a laccase from Ganoderma lucidum, and its antioxodative properties. Mol Cells. 2007, 25: 112-118.
Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropolos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, Mccombe WR, Venter JC: Complentary DNA sequencing: expressed sequence tags and human genome project. Sci. 1991, 252: 1651-1656. 10.1126/science.2047873.
La Claire JW: Analysis of expressed sequence tags from the harmful alga, Prymnesium parvum (Prymnesiophyceae, Haptophyta). Mar Biotechnol (NY). 2006, 8: 534-546. 10.1007/s10126-005-5182-2.
Suizu T, Zhou GL, Oowatari Y, Kawamukai M: Analysis of expressed sequence tags (ESTs) from Lentinula edodes. Appl Microbiol Biotechnol. 2008, 79: 461-470. 10.1007/s00253-008-1441-2.
Semova N, Storms R, John T, Gaudet P, Ulycznyj P, Min XJ, Sun J, Butler G, Tsang A: Generation, annotation, and analysis of an extensive Aspergillus niger EST collection. BMC Microbiol. 2006, 6: 7-10.1186/1471-2180-6-7.
Sacadura NT, Saville BJ: Gene expresson and EST analysis of Ustilago maydis germinating teliospores. Fungal Genet Biol. 2003, 40: 47-64. 10.1016/S1087-1845(03)00078-1.
Zhu H, Nowrousian M, Kupfer D, Colot HV, Berrocal-Tito G, Lai H, Bell-Pedersen D, Roe BA, Loros JJ, Dunlap JC: Analysis of expressed sequence tags from two starvation, time-of day-specific libraries of Neurospora crassa reveals novel clock-controlled genes. Genetics. 2001, 157: 1057-1065.
Ohlrogge J, Benning C: Unraveling plant metabolism by EST analysis. Curr Opin Plant biol. 2000, 3: 224-228.
Loo FJ, Broun van de P, Turner S, Somerville C: An oleate 12-hydroxylase from Ricinus communis L. is a fatty acyl desaturase homolog. Proc Natl Acad Sci USA. 1995, 92: 6743-6747. 10.1073/pnas.92.15.6743.
Brandle JE, Richman A, Swanson AK, Chapman BP: Leaf ESTs from Stevia rebaudiana: a resource for gene discovery in diterpene synthesis. Plant Mol Biol. 2002, 50: 613-622. 10.1023/A:1019993221986.
SuzuKi H, Achnine L, Xu R, Matsuda SPT, Dixon RA: A genomics approach to the early stages of triterpene saponin biosynthesis in Medicago truncatula. Plant J. 2002, 32: 1033-1048. 10.1046/j.1365-313X.2002.01497.x.
Jung JD, Park HW, Hahn Y, Hur CG, In DS, Chung HJ, Liu JR, Choi DW: Discovery of genes for ginsenoside biosynthesis by analysis of ginseng expressed sequence tags. Plant Cell Rep. 2003, 22: 224-230. 10.1007/s00299-003-0678-6.
Morgante M, Hanafey M, Powell W: Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet. 2002, 30: 194-200. 10.1038/ng822.
Wei JC: Fungi identification manual (in Chinese). 1979, Shanghai Scientific and Technological Press, Shanghai
Ewing B, Hillier L, Wendl MC, Green P: Base-calling of automated sequencer traces using Phred. I. accuracy assessment. Genome Res. 1998, 8: 175-185.
Ewing B, Green P: Base-calling of automated sequencer traces using Phred. II. error probabilities. Genome Res. 1998, 8: 186-194.
Laboratory of Phil Green. [http://www.phrap.org]
The UniProt-SwissProt Database. [http://www.uniprot.org/downloads]
NCBI Nr Database. [ftp://ftp.ncbi.nih.gov/blast/db/FASTA/nr.gz]
NCBI Nt Database. [ftp://ftp.ncbi.nih.gov/blast/db/FASTA/nt.gz]
Basic Local Alignment Search Tool. [ftp://ftp.ncbi.nih.gov/blast/executables/release/2.2.17/]
Simple Sequence Repeat Identification Tool (SSRIT). [http://www.gramene.org/db/markers/ssrtool]
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S: Computational and experimental analysis of microsatellites in Rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 2001, 11: 1441-1452. 10.1101/gr.184001.
Varshney RK, Graner A, Sorrells ME: Genic microsatellite markers in plants: features and applications. Trends Biotechnol. 2005, 23: 48-55. 10.1016/j.tibtech.2004.11.005.
He FM, Zhu YP, He MX, Zhang YZ: Molecular cloning and characterization of the gene encoding squalene epoxidase in Panax notoginseng. DNA Seq. 2008, 19: 270-273. 10.1080/10425170701575026.
Favre G, Ryder NS: Cloning and expression of squalene epoxidase from pathogenic yeast, Candida albicans. Gene. 1997, 189: 119-126. 10.1016/S0378-1119(96)00844-X.
Ruckenstuhl C, Leber R, Turnowsky F: Squalene epoxidase as drug target. Res Adv Antimicrob Agents Chemother. 2005, 5: 35-51.
Bun-ya M, Nishimura M, Harashima S, Oshima Y: The PH084 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol Cell Biol. 1991, 11: 3229-3238.
Schneper L, Krauss A, Miyamoto R, Fang S, Broach JR: The Ras/protein kinase A pathway acts in parallel with the Mob2/Cbk1 pathway to effect cell cycle progression and proper bud site selection. Eukaryot Cell. 2004, 3: 108-120. 10.1128/EC.3.1.108-120.2004.
Kershaw MJ, Talbot NJ: Hydrophobins and repellents: proteins with fundamental roles in fungal morphogenesis. Fungal Genet Biol. 1998, 23: 18-33. 10.1006/fgbi.1997.1022.
Ostrander DB, Gorman JA, Carman GM: Regulation of profilin localization in Saccharomyces cerevisiae by phosphoinositide metabolism. J Biol Chem. 1995, 270: 27045-27050. 10.1074/jbc.270.45.27045.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (30772735) and the National Science & Technology Pillar Program in the 11th Five-year Plan of China (2006BAI09B02-1). We thank Dr. Haibo Sun from MininGene Biotechnology (Beijing, China) for his kind help in EST analysis.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
HML analyzed the data and drafted the manuscript. CS and YL participated in the data analysis. JYS participated in the study design. JL and XWL collected tissue samples. SLC evaluated the results and revised the manuscript. All authors read and approved the final version of the manuscript.
Electronic supplementary material
13020_2009_68_MOESM1_ESM.DOC
Additional file 1: Putative functions of partial G. lucidum fruiting body ESTs. This table summarizes putative functions of partial ESTs of G. lucidum fruiting body. (DOC 86 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Luo, H., Sun, C., Song, J. et al. Generation and analysis of expressed sequence tags from a cDNA library of the fruiting body of Ganoderma lucidum. Chin Med 5, 9 (2010). https://doi.org/10.1186/1749-8546-5-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1749-8546-5-9