Abstract
Background
The sensory neurons and glia of the dorsal root ganglia (DRG) arise from neural crest cells in the developing vertebrate embryo. In mouse and chick, DRG formation is completed during embryogenesis. In contrast, zebrafish continue to add neurons and glia to the DRG into adulthood, long after neural crest migration is complete. The molecular and cellular regulation of late DRG growth in the zebrafish remains to be characterized.
Results
In the present study, we use transgenic zebrafish lines to examine neuronal addition during postembryonic DRG growth. Neuronal addition is continuous over the period of larval development. Fate-mapping experiments support the hypothesis that new neurons are added from a population of resident, neural crest-derived progenitor cells. Conditional inhibition of Notch signaling was used to assess the role of this signaling pathway in neuronal addition. An increase in the number of DRG neurons is seen when Notch signaling is inhibited during both early and late larval development.
Conclusions
Postembryonic growth of the zebrafish DRG comes about, in part, by addition of new neurons from a resident progenitor population, a process regulated by Notch signaling.
Similar content being viewed by others
Background
The neural crest, a transient population of vertebrate-specific progenitor cells, forms many disparate types of tissue after migration throughout the embryo (reviewed in [1, 2]). In the trunk, neural crest gives rise to the pigment cells of the skin and the neurons and glia of the peripheral nervous system (PNS), including the sensory neurons and glia of the dorsal root ganglia (DRG). The DRG are located in register with somites along the lateral edges of the spinal cord, and convey sensory information to the dorsal horn. The DRG are established from a population of neural crest cells that migrate along a medial pathway between somites and neural tube, to form discrete ganglia.
Mammalian and avian DRG contain three types of sensory neurons: those that relay information about touch and limb position (mechanoreceptive and proprioceptive neurons) and those that convey information about painful and irritating stimuli (nociceptive neurons). In mammals and birds, development of the DRG occurs in successive waves (reviewed in [3–5]), regulated through the activity of two neurogenin genes [6]. The first wave depends upon the activity of Neurogenin2 (neurog2), and gives rise to large-diameter mechanoreceptive and proprioceptive neurons. The second wave of neurogenesis is Neurogenin1 (neurog1)-dependent and gives rise to mechanoreceptive and nociceptive neurons. A third wave of neurogenesis arises from migration and differentiation of Krox20-expressing boundary cap cells that differentiate into nociceptive neurons [7, 8]. After neurogenesis is complete, cell death regulated by soluble neurotrophins refines the neuronal cell population to reach its final size before the end of embryogenesis.
Sensory neurogenesis in the zebrafish occurs in a somewhat different manner. Differentiation of neurons in the DRG is dependent on the activity of only one Neurogenin gene, neurog1[9], and no equivalent of neurog2 exists in the zebrafish genome. DRG initially form with only two to five neurons differentiating from the neural crest by the end of embryogenesis. Adult animals have in the range of 100 neurons [10, 11], and thus the vast majority of neurons must be added after embryogenesis. Zebrafish trigeminal sensory neurons born at different times generate different classes of neurons [12], suggesting that the postembryonic growth of the DRG might reflect a similar increase in complexity.
Here, we describe a mechanism by which zebrafish DRG neurons are added after embryogenesis. We find that neurons are added steadily throughout larval development in a process regulated by Notch signaling. In mammals, a subset of neural crest stem cells reside in an undifferentiated state in many adult tissues, including the DRG (reviewed in [13]), leading us to hypothesize that similar cells might be the source of additional neurons in zebrafish. Using in vivo lineage tracing, we find that some new neurons are generated from progenitor cells that are resident in the DRG.
Results
Neurons are continuously added to the larval dorsal root ganglia
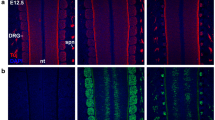
To determine whether neurons are added to the DRG in successive waves during larval development or if they are added steadily over time, we examined the total number of neuronal cells in the DRG during larval stages (six to twenty-two days postfertilization (dpf)). Larvae carrying the TgBAC(neurod:EGFP) transgene were stained with the pan-neuronal marker Elavl to label both nascent and fully differentiated DRG neurons (Figure 1A,B,C) and sorted by length in millimeters (mm) to more accurately compare fish at similar developmental stages [14]. At the earliest developmental stage examined (3.0 to 3.9 mm) there are an average of 5.7 ± 1.9 neurons per ganglion (n = 65 ganglia, 17 fish) (Figure 1C), and by the latest developmental stage examined (6 to 6.9 mm) there is an average of 30.5 ± 7.0 neurons per ganglion (n = 19 ganglia, 5 fish). The steady increase in the number of cells per ganglion demonstrates that neurons are continuously added to the DRG over larval stages (Figure 1C). To determine the pattern of neural addition during the initial formation of the DRG, we examined differentiation of sensory neurons using time-lapse imaging of the TgBAC(neurod:EGFP) line. Between 40 and 52 hours postfertilization (hpf), newly differentiating sensory neurons upregulate GFP, but do not divide (Figure 1D-D” Additional file 1). To look more closely at neuronal addition during larval development, live TgBAC(neurod:EGFP)-positive fish were examined over a period of four days when the fish were approximately 5.5 mm in length. In a representative fish, the ganglion initially contained nine neurons (Figure 1F), and during the duration of observation the ganglion added an additional four neurons (Figure 1F’,F”). Taken together, these results demonstrate that sensory neurons are added steadily to the DRG throughout the larval phase of development.
Neurons are continuously added to the larval dorsal root ganglia (DRG). (A-B) Confocal images of a TgBAC(neurod:EGFP) transgenic larva between six (A) and twenty-two dpf (B) showing DRG growth. Neurons were labeled with anti-Elavl (red) and GFP (green) antibodies. (C) Quantification (±SEM) of Elavl and GFP-positive cells per DRG in larvae staged by overall body length in millimeters (mm). Elavl-positive neurons are continuously added to the DRG as the larvae develop. Over half of the Elavl-labeled cells are also labeled with GFP (n = between five and fifteen larvae per condition, four ganglia per larva). ( -D”) Stills from a time-lapse movie showing neuron differentiation in a TgBAC(neurod:EGFP) embryo between 44 and 52 hpf. (D) At 44 hpf the DRG contain a single neuron each (blue arrowheads). (D’) By 234 minutes, neurons have been added to each ganglion (yellow arrows). (D”) At 474 minutes the ganglia contain several neurons. (E-E”) Analysis of a single ganglion in a TgBAC(neurod:EGFP) transgenic larval fish between 18 and 21 dpf. (E) At eighteen dpf, the ganglion contains nine GFP-positive cells. (E’) At 19 dpf, the ganglion contains 10 GFP-positive cells (white arrow) and by 21 dpf (E”) 12 GFP-positive cells (white arrows). During the course of imaging the larval fish grew from 5.5 mm to 5.7 mm in length. Scale bars, 20 μm. dpf, days postfertilization; hpf, hours postfertilization; GFP, green fluorescent protein.
New sensory neurons develop from progenitor cells within the dorsal root ganglia
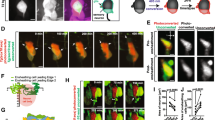
To determine which cells contribute to the expansion of sensory neurons, we analyzed the pattern of neural crest cell migration during nascent DRG formation. We used live imaging of cells in the Tg(sox10:nlsEos) transgenic line [15] to track the movement of neural crest cells. Between 18 and 32 hpf, Tg(sox10:nlsEos)-expressing neural crest cells migrate into the region of DRG formation (Figure 2 A-A”’,C; Additional file 2). Following the cessation of neural crest migration, a population of Tg(sox10:nlsEos)-positive cells remain in the DRG anlagen and form the neurons and glia of the DRG, and no additional cells are added through 56 hpf. Live imaging and cell tracking of a Tg(sox10:nlsEos)/Tg(neurod:TagRFP) embryo reveals that Eos+/RFP- cells divide and eventually give rise to RFP + DRG neurons (Figure 2B-B”’, Additional file 3). Together these results suggest that neural crest cells, as assayed by Tg(sox10:nlsEos) expression, end their migration into the DRG just prior to the onset of neurogenesis beginning at 36 hpf [9, 16, 17]. These results also demonstrate that the majority of neuronal addition occurs after neural crest migration ceases, and suggests that new neurons are derived from a population of cells within the DRG.
Neural crest migration into the dorsal root ganglia (DRG) ceases prior to neuron differentiation. (A-A”’) Confocal projections of stills from a time-lapse movie showing migrating Tg(sox10:nlsEos) + neural crest cells between 15 and 38 dpf. (A) At time zero, the region of DRG formation contains two Eos + cells. (A’) A still at 6:20 hours shows a representative neural crest cell joining the DRG anlagen (yellow arrowhead). Between 10:00 hours (A”) and 23:10 hours (A”’), no neural crest cells migrate into the DRG. (B-B”’) Time-lapse movie a Tg(sox10:nlsEos)/Tg(neurod:TagRFP) transgenic embryo between 30 and 57:10 hpf. (B) An Eos+/RFP- cell (white arrowhead) at time zero, begins to express RFP by 0:40 hours (B’) and divides at 0:50 hours (B”). (B”’) After 26.30 hours of imaging, neurons are labeled with RFP (blue arrow). (C) Quantification of Eos + cells that migrate into the DRG anlagen between 18 and 48 hpf. No new neural crest cells join the DRG after 32 hpf (n = five embryos). Scale bars, 25 μm. dpf, days postfertilization; hpf, hours postfertilization.
Previous studies analyzing embryonic and larval development in the zebrafish DRG suggested that fully differentiated neurons are a source of newly added neurons as assayed by either bromodeoxyuridine (BrdU) incorporation or anti-phosphohistone H3 (pH3) labeling [10, 18]. To address which cells divide within the DRG in more detail, larval fish were pulsed with the thymidine analog BrdU to mark cells in S-phase and/or labeled with anti-pH3 antibody to mark cells in M-phase, and labeling was carefully examined by confocal microscopy. In contrast to previous research, we did not find evidence of large-scale neuronal proliferation in the DRG. In five dpf larvae that were labeled with anti-pH3 antibody, only a small percentage of DRG had pH3+ cells in proximity to Elavl + neurons and no overlap in labeling was present (Figure 3A; 19 of 256 DRG (6%) had one or more associated pH3+ cells, n = 10 larvae). To further assess the role that proliferation plays during neuronal addition to the DRG, we preformed both acute and pulse-chase analyses of BrdU incorporation. In larvae that were incubated in BrdU immediately prior to fixation at five dpf, the vast majority of BrdU + cells associated with the DRG did not express Elavl, though a small subset of BrdU+/Elavl + cells was identified (2/204; Figure 3B; Table 1). Larvae that were exposed to a pulse of BrdU at two dpf and BrdU incorporation was analyzed at five dpf showed a small, but not significant increase in the number of BrdU+/Elavl + cells (Figure 3C; Table 1). Incorporation was also assessed in DRG of late-stage larval fish (25 dpf) two hours after BrdU injection. The DRG at 25 dpf contained both BrdU + cells (an average of 6.47 ± 0.54 per ganglion) and pH3+ cells (an average of 0.23 ± 0.08 per ganglion), but careful analysis of individual confocal slices revealed that in no cases were these cells also Elavl + (there were an average of 19.42 ± 1.28 Elavl + per ganglion; ± SEM; n = 15 fish) (Figure 3D; Additional file 4). These data suggest that non-neuronal cells associated with the DRG are proliferating and may act as precursor cells that give rise to new neurons as the DRG expands.
Non-neuronal cells associated with the dorsal root ganglia (DRG) proliferate. (A) Confocal projection of a ganglion at five dpf, showing sensory neurons labeled with Elavl (green) and an associated cell labeled with pH3 (red; blue arrow). Nuclei were labeled with DAPI (blue). (B-C) Ganglia of larvae at five dpf following BrdU incorporation (red). Neurons are labeled with Elavl (red) and nuclei are labeled with DAPI (blue). (B) Following a BrdU pulse at five dpf, cells associated with the DRG incorporate BrdU (yellow arrowhead), but neurons do not. (C) In a larva that was pulsed with BrdU at two dpf and chased to five dpf, a BrdU + neuron is located in the DRG (white arrowhead). (D) A ganglion in a 25 dpf, 6.25 mm larval fish exposed to a pulse of BrdU at 25 dpf, shows BrdU + cells (red; yellow arrowhead) and pH3 labeled cells (blue; blue arrow) are intermingled among Elavl + neurons (green), no neurons are labeled with BrdU or pH3. Dashed lines indicate levels of XZ and YX insets. Scale bars, 20 μm. BrdU, bromodeoxyuridine; DAPI, 4',6-diamidino-2-phenylindole; dpf, days postfertilization. pH3, phosphohistone H3.
To specifically address this hypothesis, we used the Tg(sox10:nlsEos) transgenic line to label and follow cells in vivo. The sox10 promoter construct directs expression in migrating neural crest cells, in neural crest-derived glia and in cells fated to differentiate as sensory neurons [18]. The Eos protein undergoes irreversible photoconversion when exposed to 405 nm light [19], and the nuclear localization signal allows unambiguous labeling of single cells. Based on our findings that neural crest migration is complete prior to neurogenesis (Figure 2) and the findings of Carney et al. [18], we propose that the Tg(sox10:nlsEos) positive cells present in the DRG by four dpf includes non-neuronal cells comprised of satellite glial cells and progenitor cells; at present, there are no definitive zebrafish markers that allow us to distinguish between these cell types. Note also that the nls-Eos protein continues to be expressed in Elavl + cells, presumably because this protein is stable and persists, as Sox10 protein is downregulated in differentiated neurons [9]. At four dpf, the DRG contains an average six ± 0.19 Tg(sox10:nlsEos)-expressing cells and 2.8 ± 0.13 Elavl + cells (Figure 4A; ± SEM; n = five larvae). By six dpf, the DRG has expanded to contain an average eight ± 0.33 Tg(sox10:nlsEos)-expressing cells and 3.1 ± 0.16 Elavl + cells (Figure 4B; ± SEM; n = 13 larvae).
Dividing progenitors within the larval dorsal root ganglia (DRG). (A-B) Confocal projections of ganglia-containing cells that express the Tg(sox10:nlsEos) transgene (green) and neurons labeled with Elavl (red). At four dpf (A) and six dpf (B) Tg(sox10:nlsEos) labels both neurons (yellow arrowheads) and non-neuronal cells. (C-E) Tg(sox10:nlsEos) cells were photoconverted by brief exposure to 405 nm light at four dpf, resulting in red fluorescence (white arrow). (C’-E’) At six dpf, larvae were labeled with Elavl (blue) and analyzed by confocal microscopy. Shown are examples of cells that divided and gave rise to two sensory neurons (SN-SN; yellow arrowheads) (C-C’), two non-neuronal cells (NN-NN; white arrowheads) (D-D’) or one sensory neuron and one non-neuronal cell (E-E’). Scale bar, 20 μm. dpf, days postfertilization.
To identify the source of new neurons, we used photoconversion of Eos at four dpf to label cells associated with the DRG, and assessed their phenotypes at six dpf. Neurons and non-neuronal cells were distinguished by expression of Elavl. About two-thirds of labeled cells did not divide (Table 2). Of these, one-third was identified as sensory neurons, including some Eos + cells that were presumably already Elavl + at the time of photoconversion at four dpf. In some cases, photoconverted cells divided to generate two sensory neurons (Figure 4C,C’) or non-neuronal cells (Figure 4D,D’). Further, a few cells underwent asymmetric division to give rise to both sensory neurons and non-neuronal cells (Figure 4E,E’). These results demonstrate that some neurons are derived from dividing potential progenitor cells associated with the DRG.
delta and notch are expressed in cells associated with dorsal root ganglia neurons
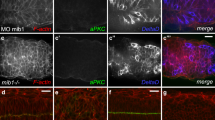
We next sought to uncover the mechanism underlying the differentiation of DRG neurons during larval development. The Delta/Notch signaling pathway is a central player in the establishment of neuronal fates in many systems (see [20, 21] for review). Delta/Notch signaling has been shown to perform an important role in specification of neural crest and Rohon-Beard sensory neurons in the zebrafish [17, 22], and specification of DRG neurons and glia in chick [23]. To determine whether the Delta/Notch signaling pathway may act to regulate the specification of zebrafish DRG sensory neurons from a progenitor population by a similar mechanism, we used fluorescent RNA in situ hybridization to identify the expression patterns of notch1a delta A and deltaD genes in the DRG at 48 hpf. We found that expression of notch1a deltaA and deltaD is seen in cells neighboring both Tg(neurog1:EGFP)-expressing cells (Figure 5A,-C) and Elavl + neurons (Figure 5D-F). These results suggest that non-neuronal cells associated with DRG might use the differential expression of delta and notch mRNAs to regulate progenitor cell differentiation.
delta and notch mRNAs are expressed in cells associated with dorsal root ganglia (DRG) neurons. (A-C) Confocal images of notch1a, deltaA and deltaD fluorescent RNA in situ hybridization in 48 hpf Tg(neurog1:EGFP) transgenic embryos. (A) Expression of notch1a (red, dashed line) is found in a cell that neighbors GFP + DRG neuron (green). Both deltaA (B) and deltaD (C) are expressed in cells surrounding GFP + neurons. ( -F) Comparison of anti-Elavl labeling and notch1a, deltaA and deltaD RNA expression at 48 hpf. (D) A cell expressing notch1a (red) is seen adjacent to an Elavl-positive neuron (green). (E) deltaA is expressed in cells surrounding Elavl-positive neurons, as is deltaD (F). Scale bars, 20 μm. hpf, hours postfertilization.
Inhibition of Notch signaling increases neuron number during larval dorsal root ganglia development
Genetic disruption of Notch signaling results in loss of neural crest induction in developing zebrafish embryos [17, 22], thus preventing the analysis of the requirement of this pathway at subsequent steps of development. To test the role of the Delta/Notch signaling pathway in specification of sensory neurons in the zebrafish DRG, we employed two methods of conditionally inhibiting Notch signaling. The gamma-secretase mediated cleavage of the Notch intracellular domain (ICD) was blocked with the pharmacological inhibitor DAPT, which has been shown to phenocopy Notch signaling mutants in zebrafish [24]. To confirm the efficacy of Notch inhibition by DAPT, we used transgenic line, Tg(hsp70l:XdnSu(H)myc), where a dominant-negative form of Suppressor of Hairless [25] is expressed following induction by heat-shock [26]. To determine whether the Delta/Notch signaling pathway acts after neural crest migration is complete, we continuously disrupted Notch signaling in Tg(neurog1:EGFP) larvae between two and five dpf. Treatment with DAPT during this period resulted in a significant increase in the number of DRG neurons as compared to DMSO-treated fish (Figure 6A,B,E). A similar result was seen using Tg(hsp70l:XdnSu(H)myc) larvae that were heat-shocked every 12 hours between two and five dpf (Figure 6C,D,E). Notch inhibition in a second transgenic line, TgBAC(neurod:EGFP), resulted in a comparable significant increase in the total number of GFP-positive and Elavl-positive neurons (Figure 6F).
Notch inhibition during larval development leads to an increase in dorsal root ganglia (DRG) neurons. (A-B) Confocal projections of Tg(neurog1:EGFP) DRG neurons labeled with anti-GFP (green) and Elavl (red) from larvae treated with DMSO or DAPT between two and five dpf. At five dpf, DRG neurons are labeled by Elavl (yellow arrowhead) and a subset also express GFP (white arrow) in both DMSO (A) and DAPT (B) treated larvae. (C-D) Confocal images of Tg(hsp70l:XdnSu(H)myc); Tg(neurog1:EGFP) larvae following heat-shock (HS) beginning at two dpf. In control (C) and transgenic (D) larvae at five dpf after HS, DRG neurons were labeled with GFP (green) and Elavl (red). Larvae carrying the Tg(hsp70l:XdnSu(H)myc) transgene were identified by ectopic myc labeling (blue, white asterisk) following HS. (E) Quantification of DRG neurons following Notch inhibition with DAPT or HS in Tg(hsp70 DN (Su(H) myc) embryos from two to five dpf. There is a significant increase in the number of DRG neurons in DAPT-treated and myc-positive larvae as compared to control larvae. (n = 6 for DMSO- or DAPT-treated larvae, n = 10 for myc-negative larvae and n = 13 for myc-positive larvae, five ganglia per larvae, *P <0.03, Student’s t test). (F) Quantification of DRG neurons in TgBAC(neurod:EGFP)-positive DRG following Notch inhibition with DAPT from two to five dpf. There is a significant increase in the number of DAPT-positive DRG neurons as compared to control embryos. (n = 9 for DMSO or n = 8 for DAPT-treated larvae, five ganglia per larvae, *P <0.005, Student’s t test). Scale bars, 20 μm. DAPT, N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester; DMSO, dimethyl sulfoxide; dpf, days postfertilization GFP, green fluorescent protein.
To determine if proliferation is required for the increase in DRG neurons following Notch inhibition, we used acute or pulse-chase BrdU incorporation following the strategy described above. For the pulse-chase paradigm, larvae were treated with BrdU at two dpf and then incubated in DMSO or DAPT from two to five dpf. For acute BrdU experiments, larvae were exposed to DMSO or DAPT from two to five dpf and then treated with BrdU at five dpf just prior to fixation. Both DAPT treatment conditions showed a significant increase in the number of Elavl + neurons as compared to DMSO controls (Table 1). Following DAPT treatment, no significant increase was seen in BrdU + cells or BrdU+/Elavl + cells in either condition when compared to DMSO controls. These results suggest that blocking Notch signaling results in addition of DRG neurons primarily by promoting differentiation rather than increasing proliferation of progenitor cells.
Addition of neurons in late-stage larval dorsal root ganglia is sensitive to Notch inhibition
Zebrafish begin to transition from their larval-to-adult form during a metamorphic period that occurs between approximately 14 and 28 dpf based on changes in pigment pattern, physiology and behavior [14]. To determine whether the Delta/Notch signaling pathway continues to regulate neurogenesis during later stages, late larval fish were treated with DMSO or DAPT between 20 and 25 dpf. Both control and DAPT-treated larvae developed to an average of 6 mm in length. Despite being of similar developmental stage, the DRG of DAPT-treated fish contained significantly more neurons then DMSO-treated controls (an average of 29.8 and 21.3 respectively; n = 10 larvae per treatment group; P = 0.002; Figure 7A-C). Therefore, Notch signaling plays a role throughout larval development to regulate the specification of DRG neurons.
Excess neurons differentiate in late larval dorsal root ganglia (DRG) following Notch inhibition. (A-B) Confocal images of TgBAC(neurod:EGFP) transgenic fish treated with DMSO or DAPT between 20 and 25 dpf. DRG neurons were labeled with anti-GFP antibody (green) and Elavl (red). At 25 dpf, the DRG of a 6.3 mm DMSO-treated fish (A) and a 6.1 mm DAPT-treated fish (B) contain neurons that are labeled with Elavl, GFP/Elavl or GFP antibody alone. (C) Quantification of cells in the DRG. Following DAPT treatment, there is a significant increase in the total number of GFP + and Elavl + cells in the DRG as compared to controls. (n = 10 fish per condition, four ganglia per fish, *P <0.002, Student’s t test). Scale bars, 20 μm. DAPT, N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester; DMSO, dimethyl sulfoxide; dpf, days postfertilization GFP, green fluorescent protein.
Discussion
Zebrafish, like other aquatic vertebrates, undergo substantial growth after embryogenesis is complete, including in the size and complexity of their nervous system. We demonstrate the addition of new neurons to the zebrafish DRG is continuous during postembryonic (larval) growth. Similar increases in DRG neuron number have been reported during amphibian metamorphosis [27]. By contrast, growth of DRG in avian and mammalian species occurs during embryogenesis in overlapping waves that produce neurons of distinct function [6, 28–32]. It remains to be determined whether distinct neuronal classes are added to the zebrafish DRG at specific developmental stages. While postembryonic growth of DRG in mammals has been controversial, addition of neurons in other mammalian PNS structures has been well documented [33, 34].
Our lineage tracing experiments demonstrate that progenitors that reside among the Sox10-positive non-neuronal cells of the DRG are one source of new neurons during postembryonic growth. These results are consistent with a recent study demonstrating the addition of new DRG neurons from peripherally located precursors during avian embryogenesis [30]. In zebrafish, this process appears to continue long after embryogenesis has ended. We demonstrate that zebrafish progenitor cells are associated with the zebrafish DRG after neural crest migration is complete, although these cells share Sox10 expression with the neural crest cells from which they are derived. Previous studies in mammals have suggested continued addition of neurons during late embryonic development from other sources, including neural crest-derived boundary cap cells found at the dorsal and ventral roots [35–37]. Boundary cap cells can form a number of late-developing sensory neurons [7, 8] and also acquire glial characteristics [38, 39]. We believe that the progenitors we have identified in zebrafish are distinct from boundary cap cells, as they are located in positions distinct from the dorsal and ventral roots, and do not express Krox20/egr2 mRNA (data not shown).
While sensory neurons are generally thought to be postmitotic, two previous studies have suggested that zebrafish DRG Elav + cells divide to give rise to new neurons [10, 18]. These results are in contrast to our findings and to those in other systems [30]. We performed careful three-dimensional analysis to distinguish labeled cells, and note that while we have identified a very small number of Elav + cells that have incorporated BrdU, we do not find cells that are labeled with pH3. Furthermore, by time-lapse analysis we find that differentiated neurons do not divide, but rather arise de novo. We conclude that dividing Elav + cells are not a major source of DRG neurons in zebrafish.
We have not resolved whether DRG sensory neuron progenitors are bona fide glial cells, or whether they are a distinct population. Nervous system stem cells are now well established to have glial cell characteristics (reviewed in [40, 41]), including those found in the PNS. We identified potential DRG progenitors by continued expression of the Tg(sox10:nlsEos) transgene and their positions adjacent to DRG neurons. Whether these cells acquire characteristics of satellite glia, which surround sensory neuron cell bodies and alter neuronal function [42, 43], will require further study.
Our data suggests that Notch signaling regulates sensory neuron addition. We found that non-neuronal cells associated with differentiated DRG neurons express the Notch ligands deltaA and deltaD, as well as the Notch receptor notch1a. Inhibition of Notch signaling induces differentiation of DRG neurons in early- and late-stage larval fish. Our results are consistent with previous reports of an increase in DRG neuron number in zebrafish notch1a mutants [44]. Recent reports have also demonstrated that loss of Notch signaling increases the initial differentiation of DRG neurons from neural crest [45, 46]. Our results suggest that Notch signaling normally prevents differentiation of neuronal progenitors. The expression of both ligand and receptor in subsets of cells adjacent to DRG neurons suggests a model where progenitor cells may act as an equivalence group: as cells begin to differentiate they inhibit their neighbors from doing so. One test of equivalence, whether cells are replaced by their neighbors after genetic or mechanical ablation, awaits future study. The methods we have used to block Notch signaling will act ubiquitously throughout the embryo, leaving open the possibility that effects are nonautonomous. In this scenario, blocking Notch would affect another cell type that would subsequently influence DRG development. Further experiments are needed to determine if Notch signaling acts cell autonomously within DRG progenitors.
Multiple roles for Notch signaling in neural crest development and differentiation have been previously identified, suggesting that this signaling pathway functions in a context-dependent manner. Notch signaling plays a role in the establishment of neural crest [17, 22, 47], and in the development of neural crest-derived cartilage and heart tissue [48]. Multiple roles for Notch signaling have also been described for glial cell specification and differentiation [49–52]. Here we describe a later role for Notch in regulating the differentiation of neuronal progenitors during postembryonic phases of zebrafish DRG development. Similar roles have been previously described for Notch signaling in avian sensory and sympathetic progenitors [53] and mouse enteric progenitors [54]. While the different roles for Notch signaling might be due to context or timing, the relative level of pathway stimulation may also have an effect [55]. In addition, oscillating expression of Notch signaling components alters the effects of this signaling pathway in the embryonic brain [56]. In cultures of neural crest stem cells, reduced levels of Notch signaling inhibit neurogenesis but do not promote gliogenesis, thus promoting self-renewal and maintaining pluripotency [57].
The presence of latent progenitors for postembryonic DRG growth is intriguing, and suggests a possible source of cells for regeneration. In mammals, neural crest-derived resident stem cells have been documented in a number of adult tissues, including amongst the enteric nervous system, heart, peripheral nerves and skin [58–64]. Neural crest stem or progenitor cells have also been isolated from embryonic and adult mammalian DRG [61, 65–70]. These cells may be involved in replacement or repair instead of growth, the possible evolutionary origin of their function.
Conclusions
Our work demonstrates the orderly addition of sensory neurons to the zebrafish DRG in the weeks after embryogenesis is complete, long after neural crest migration is complete. Neurons arise from dividing latent progenitor cells associated with the DRG that are amongst the satellite glial population. Progenitor differentiation is regulated by Notch signaling, throughout the period of larval development. The work provides insight into sensory neurogenesis in animals that undergo dramatic postembryonic growth.
Methods
Transgenic zebrafish lines
Larval and adult zebrafish were maintained at 28.5°C in a 14 hour/10 hour light–dark cycle with twice daily feedings. Embryos were generated from natural crosses between adults and staged in hours post fertilization (hpf) as described by [71]. Larval fish were staged in days postfertilization (dpf) and standard length [14]. The transgenic lines used were: Tg(neurog1:EGFP)w61 described in [9]; TgBAC(neurod:EGFP)nl1 described in [72], Tg(sox10:nlsEos)w18 described in [15] and Tg(hsp70:XdnSu(H)myc)vu21 described in [26]. Construction of the Tg(neurod:TagRFP)w69 transgenic line is described below. All work was approved by the University of Washington Institutional Animal Use and Care Committee.
Construction of Tg(neurod:TagRFP)
The Tg(neurod:TagRFP)w69 transgenic line was constructed using the Multisite Gateway system [73]. The p5E-neuroD construct was generated using a 5 kb region of the neurod 5′ promoter ([72]; a gift from Teresa Nicolson). The p5E-neuroD construct was recombined with pME-TagRFP (a gift from Chi-Bin Chien) and p3E polyA. The construct was injected into one-cell embryos with Tol2 polymerase [74]. Injected embryos were raised to sexual maturity and screened by pair-wise crosses to obtain germ line transgenic fish that generated the expected expression pattern. The final construct was injected into one-cell zygotes, which were then raised to maturity and screened for progeny that carried the Tg(neurod:TagRFP) transgene in DRG neurons amongst other cells.
Whole-mount immunohistochemistry and RNA in situ hybridization
For immunohistochemistry, larvae were collected at the stages indicated, euthanized in MS-222 (Sigma-Aldrich, St Louis, MO, USA); 10 mg/ml in buffered embryo medium) and fixed in 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS) for two hours at room temperature. Antibody labeling was carried out as previously described [75]. In brief, embryos were washed in PBS with 0.1% TritonX-100 (PBT), and blocked with the addition of 2% goat serum. Prior to blocking, fish were permeablized with three one-hour water washes. Fish were incubated in primary antibodies diluted in blocking solution overnight at room temperature (RT). Primary antibodies used were anti-GFP (1:700; rabbit or mouse anti-GFP; Invitrogen, Carlsbad, CA, USA), anti-Elavl (1:700; monoclonal antibody (mAB) 16A11; also called anti-HuC/D; [76]; Invitrogen); anti-myc tag mAB (1:500, Cell Signaling Technology, Beverly, MA, USA), anti-5-bromo-2-deoxyuridine (rat anti-BrdU; 1:100; Abcam, Cambridge, UK) and anti-phosphohistone H3 (rabbit anti-pH3; 1:100, Cell Signaling Technology). Fish were incubated in Alexa-488, Alexa-568 or Alexa-647 conjugated secondary antibodies (Invitrogen) overnight at room temperature, rinsed in PBT and then stored in 50% glycerol/PBS for imaging. Nuclei were visualized by DAPI labeling (Invitrogen).
Fluorescent RNA in situ hybridization was performed as described [77]. Fixed larvae were made permeable for 20 minutes at RT using 10 μg/ml Proteinase K (Sigma-Aldrich) in PBT and then refixed for 15 minutes in 4% PFA. All hybridizations were carried out at 55°C. Digoxygenin-labeled antisense probes were generated using the following restriction enzyme and polymerases: XbaI/T7 for notch1a[78]; EcoR1/T7 for deltaA and deltaD[79, 80]. Following in situ hybridization, larvae were processed for immunohistochemistry as described above. Larvae were then stored in 50% glycerol/PBS for imaging.
Inhibition of Notch signaling
The pharmacological inhibitor of gamma-secretase activity N-N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT; [24]; Sigma-Aldrich) was used to conditionally inhibit Notch signaling. DAPT was dissolved in dimethylsulfoxide (DMSO) at 10 mM and a working stock was diluted to 100 μM in embryo medium (EM) as previously described [81]. Control fish were mock-treated with 1% DMSO in EM, for the same time course as the DAPT-treated fish. Zebrafish larvae were examined for the role of Notch signaling at two time points, either during early larval development, between two and five dpf, or during late larval development, between twenty and twenty-five dpf. Fish were treated continuously with DAPT or DMSO and were fed daily for the duration of the treatment. Following fixation, fish were processed for immunohistochemistry.
To corroborate the results seen with DAPT treatment, a heat-shock inducible dominant negative Suppressor of Hairless transgenic, Tg(hsp70l:XdnSu(H)myc), was used [26]. Larvae were heat-shocked at 40°C for one hour twice daily between two and five dpf, fixed in 4% PFA, and processed for immunohistochemistry. Activation of the transgene was confirmed by the presence of myc labeling with an anti-myc antibody (Invitrogen).
Cell cycle analysis
To assess cell cycle in the DRG, we used 5-bromo-2-deoxyuridine (BrdU) incorporation to label cells in S-phase and anti-phosphohistone H3 (pH3) antibody to label cells in M-phase [82]. In larvae, BrdU incorporation was carried out using a modified established protocol [83] Larvae were incubated in 10 mM BrdU dissolved in EM and 10% DMSO for 30 minutes at 4°C followed by one hour at 28.5°C. For pulse-chase experiments, larvae were exposed to BrdU between 50 and 51.5 hpf, transferred to DAPT or DMSO until five dpf and then fixed and processed. For pulse experiments, larvae that had been treated with DAPT or DMSO between two and five dpf, were exposed to BrdU at five dpf using the above protocol and then fixed and processed for BrdU detection. For BrdU incorporation in older larval fish, animals at 25 dpf were anesthetized in MS-222 and injected intraperitoneally using a glass capillary needle with 10 nl of a 10 mM BrdU in buffered Hank’s solution (protocol modified from [84]), incubated in EM at 28.5°C for two hours and then fixed for processing. BrdU detection was carried out using a previously describe protocol [85] Following BrdU detection, fish were immunolabeled with anti-Elavl and anti-pH3 antibodies and stored in 50% glycerol/PBS for imaging.
In vivo fate mapping
Homozygous larvae carrying the Tg(sox10:nlsEos) transgene were raised to four dpf, anesthetized with MS-222, and mounted in 0.8% agarose on a glass-bottomed cell culture dish. Cells surrounding the DRG were exposed to 405 nm light on an Olympus FV1000 confocal microscope (Olympus, Tokyo, Japan) to convert individual green nuclei to red. Following photoconversion, larvae were released from the agarose, and incubated in either 100 uM DAPT or DMSO as a control. At six dpf, larvae were fixed in 4% paraformaldehyde for two hours at room temperature and labeled with anti-ElavI antibody, mounted in Vectashield (Vector Laboratories, Burlingame, CA, USA) and imaged.
Data collection and time-lapse imaging
Prior to imaging, fish were mounted on bridged cover slips in 50% glycerol/PBS. Images and cell counts were obtained using a Zeiss LSM-5.0 Pascal confocal microscope (Carl Zeiss AG., Oberkochen, Germany), a Zeiss Axioplan 2 microscope and a Spot CCD camera (Diagnostic Instruments, Palo Alto, CA, USA), an Olympus FV1000 confocal microscope, or a 3I Marianas spinning disk microscope (Intelligent Imaging Innovations, Inc., Denver, CO, USA). Whole images were processed using ImageJ software (rsbweb.nih.gov/ij/index.html) and adjusted for brightness and contrast using Adobe Photoshop. For most individual cell counts, the five rostral-most DRG were analyzed in each condition. Non-neuronal cells were defined as associated with the DRG if they were in direct contact with labeled DRG neuronal cell bodies. Statistical significance was determined using Microsoft Excel software (Microsoft, Seattle, WA, USA) or GraphPad Prism version 5.0d software (Graphpad Software, San Diego, CA, USA).
For live imaging of neural crest migration and initial differentiation of neurons, embryos were anesthetized in MS-222, then embedded in 1.2% low melt agarose (Sigma-Aldrich) in EM. Cells were imaged every 10 minutes with a 20x water lens. In some cases stacks were assembled using Slidebook. Cells were tracked manually using ImageJ software. To image neuronal addition in larval fish, animal were anesthetized and embedded in 1.5% low-melt agarose and imaged using a 40x dipping lens. Following imaging, fish were released from the agarose and revived. Images were collected daily over a four-day time course.
Abbreviations
- BrdU:
-
Bromodeoxyuridine
- DAPI 4′:
-
6-diamidino-2-phenylindole
- DAPTN-[N-(3:
-
5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester
- DMSO:
-
Dimethyl sulfoxide
- DRG:
-
Dorsal root ganglion
- dpf:
-
Days postfertilization
- EGFP:
-
Enhanced green fluorescent protein
- EM:
-
Embryo medium
- GFP:
-
Green fluorescent protein
- hpf:
-
Hours postfertilization
- IVD:
-
Intracellular domain
- PBS:
-
Phosphate buffered saline
- PFA:
-
Paraformaldehyde
- pH3:
-
Phosphohistone H3
- RT:
-
Room temperature
- TagRFP:
-
Tag red fluorescent protein.
References
Hall BK, Hörstadius S: The neural crest: including a facsimile reprint of The neural crest by Sven Horstadius. 1988, Oxford University Press, London; New York
Le Douarin N, Kalcheim C: The neural crest. 1999, Cambridge University Press, Cambridge, UK; New York, NY, USA
Marmigère F, Ernfors P: Specification and connectivity of neuronal subtypes in the sensory lineage. Nat Rev Neurosci. 2007, 8: 114-127. 10.1038/nrn2057.
Pavan WJ, Raible DW: Specification of neural crest into sensory neuron and melanocyte lineages. Dev Biol. 2012, 366: 55-63. 10.1016/j.ydbio.2012.02.038.
Raible DW, Ungos JM: Specification of sensory neuron cell fate from the neural crest. Adv Exp Med Biol. 2006, 589: 170-180. 10.1007/978-0-387-46954-6_10.
Ma Q, Fode C, Guillemot F, Anderson DJ: Neurogenin1 and neurogenin2 control two distinct waves of neurogenesis in developing dorsal root ganglia. Genes Dev. 1999, 13: 1717-1728. 10.1101/gad.13.13.1717.
Hjerling-Leffler J, Marmigère F, Heglind M, Cederberg A, Koltzenburg M, Enerbäck S, Ernfors P: The boundary cap: a source of neural crest stem cells that generate multiple sensory neuron subtypes. Development. 2005, 132: 2623-2632. 10.1242/dev.01852.
Maro GS, Vermeren M, Voiculescu O, Melton L, Cohen J, Charnay P, Topilko P: Neural crest boundary cap cells constitute a source of neuronal and glial cells of the PNS. Nat Neurosci. 2004, 7: 930-938. 10.1038/nn1299.
McGraw HF, Nechiporuk A, Raible DW: Zebrafish dorsal root ganglia neural precursor cells adopt a glial fate in the absence of neurogenin1. J Neurosci. 2008, 28: 12558-12569. 10.1523/JNEUROSCI.2079-08.2008.
An M, Luo R, Henion PD: Differentiation and maturation of zebrafish dorsal root and sympathetic ganglion neurons. J Comp Neurol. 2002, 446: 267-275. 10.1002/cne.10214.
Weis JS: Analysis of the development of the nervous system of the zebrafish, Brachydanio rerio. J Embryol Exp Morphol. 1968, 19: 109-119.
Caron SJ, Prober D, Choy M, Schier AF: In vivo birthdating by BAPTISM reveals that trigeminal sensory neuron diversity depends on early neurogenesis. Development. 2008, 135: 3259-3269. 10.1242/dev.023200.
Shakova O, Sommer L: Neural crest-derived stem cells. StemBook. Edited by: Girard P. 2010, Harvard Stem Cell Institute, Cambridge
Parichy DM, Elizondo MR, Mills MG, Gordon TN, Engeszer RE: Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Dev Dyn. 2009, 238: 2975-3015. 10.1002/dvdy.22113.
Prendergast A, Limbo TH, Swarts T, Ungos JM, McGraw HF, Krispin S, Weinstein BM, Raible DW: The metalloproteinase inhibitor RECK is essential for zebrafish DRG development. Development. 2012, 139: 1141-1152. 10.1242/dev.072439.
Andermann P, Ungos J, Raible DW: Neurogenin1 defines zebrafish cranial sensory ganglia precursors. Dev Biol. 2002, 251: 45-58. 10.1006/dbio.2002.0820.
Cornell RA, Eisen JS: Delta/Notch signaling promotes formation of zebrafish neural crest by repressing Neurogenin 1 function. Development. 2002, 129: 2639-2648.
Carney TJ, Dutton KA, Greenhill E, Delfino-Machín M, Dufourcq P, Blader P, Kelsh RN: A direct role for Sox10 in specification of neural crest-derived sensory neurons. Development. 2006, 133: 4619-4630. 10.1242/dev.02668.
Wiedenmann J, Ivanchenko S, Oswald F, Schmitt F, Röcker C, Salih A, Spindler KD, Nienhaus GU: EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc Natl Acad Sci USA. 2004, 101: 15905-15910. 10.1073/pnas.0403668101.
Corbin JG, Gaiano N, Juliano SL, Poluch S, Stancik E, Haydar TF: Regulation of neural progenitor cell development in the nervous system. J Neurochem. 2008, 106: 2272-2287. 10.1111/j.1471-4159.2008.05522.x.
Louvi A, Artavanis-Tsakonas S: Notch signaling in vertebrate neural development. Nat Rev Neurosci. 2006, 7: 93-102. 10.1038/nrn1847.
Cornell RA, Eisen JS: Delta signaling mediates segregation of neural crest and spinal sensory neurons from zebrafish lateral neural plate. Development. 2000, 127: 2873-2882.
Wakamatsu Y, Maynard TM, Weston JA: Fate determination of neural crest cells by NOTCH-mediated lateral inhibition and asymmetrical cell division during gangliogenesis. Development. 2000, 127: 2811-2821.
Geling A, Steiner H, Willem M, Bally-Cuif L, Haass C: A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep. 2002, 3: 688-694. 10.1093/embo-reports/kvf124.
Wettstein DA, Turner DL, Kintner C: The Xenopus homolog of Drosophila Suppressor of Hairless mediates Notch signaling during primary neurogenesis. Development. 1997, 124: 693-702.
Latimer AJ, Appel B: Notch signaling regulates midline cell specification and proliferation in zebrafish. Dev Biol. 2006, 298: 392-402. 10.1016/j.ydbio.2006.05.039.
Farel PB, McIlwain DL: Neuron addition and enlargement in juvenile and adult animals. Brain Res Bull. 2000, 53: 537-546. 10.1016/S0361-9230(00)00387-7.
Carr VM, Simpson SB: Proliferative and degenerative events in the early development of chick dorsal root ganglia. I. Normal development. J Comp Neurol. 1978, 182: 727-739. 10.1002/cne.901820410.
George L, Chaverra M, Todd V, Lansford R, Lefcort F: Nociceptive sensory neurons derive from contralaterally migrating, fate-restricted neural crest cells. Nat Neurosci. 2007, 10: 1287-1293. 10.1038/nn1962.
George L, Kasemeier-Kulesa J, Nelson BR, Koyano-Nakagawa N, Lefcort F: Patterned assembly and neurogenesis in the chick dorsal root ganglion. J Comp Neurol. 2010, 518: 405-422. 10.1002/cne.22248.
Kitao Y, Robertson B, Kudo M, Grant G: Neurogenesis of subpopulations of rat lumbar dorsal root ganglion neurons including neurons projecting to the dorsal column nuclei. J Comp Neurol. 1996, 371: 249-257. 10.1002/(SICI)1096-9861(19960722)371:2<249::AID-CNE5>3.0.CO;2-2.
Lawson SN, Biscoe TJ: Development of mouse dorsal root ganglia: an autoradiographic and quantitative study. J Neurocytol. 1979, 8: 265-274. 10.1007/BF01236122.
Lagares A, Li HY, Zhou XF, Avendaño C: Primary sensory neuron addition in the adult rat trigeminal ganglion: evidence for neural crest glio-neuronal precursor maturation. J Neurosci. 2007, 27: 7939-7953. 10.1523/JNEUROSCI.1203-07.2007.
Pardal R, Ortega-Sáenz P, Durán R, López-Barneo J: Glia-like stem cells sustain physiologic neurogenesis in the adult mammalian carotid body. Cell. 2007, 131: 364-377. 10.1016/j.cell.2007.07.043.
Altman J, Bayer SA: The development of the rat spinal cord. Adv Anat Embryol Cell Biol. 1984, 85: 1-164. 10.1007/978-3-642-69537-7_1.
Golding JP, Cohen J: Border controls at the mammalian spinal cord: late-surviving neural crest boundary cap cells at dorsal root entry sites may regulate sensory afferent ingrowth and entry zone morphogenesis. Mol Cell Neurosci. 1997, 9: 381-396. 10.1006/mcne.1997.0647.
Niederländer C, Lumsden A: Late emigrating neural crest cells migrate specifically to the exit points of cranial branchiomotor nerves. Development. 1996, 122: 2367-2374.
Aquino JB, Hjerling-Leffler J, Koltzenburg M, Edlund T, Villar MJ, Ernfors P: In vitro and in vivo differentiation of boundary cap neural crest stem cells into mature Schwann cells. Exp Neurol. 2006, 198: 438-449. 10.1016/j.expneurol.2005.12.015.
Coulpier F, Le Crom S, Maro GS, Manent J, Giovannini M, Maciorowski Z, Fischer A, Gessler M, Charnay P, Topilko P: Novel features of boundary cap cells revealed by the analysis of newly identified molecular markers. Glia. 2009, 57: 1450-1457. 10.1002/glia.20862.
Costa MR, Götz M, Berninger B: What determines neurogenic competence in glia?. Brain Res Rev. 2010, 63: 47-59. 10.1016/j.brainresrev.2010.01.002.
Kriegstein A, Alvarez-Buylla A: The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009, 32: 149-184. 10.1146/annurev.neuro.051508.135600.
Hanani M: Satellite glial cells in sensory ganglia: from form to function. Brain Res Brain Res Rev. 2005, 48: 457-476.
Scholz J, Woolf CJ: The neuropathic pain triad: neurons, immune cells and glia. Nat Neurosci. 2007, 10: 1361-1368. 10.1038/nn1992.
Gray M, Moens CB, Amacher SL, Eisen JS, Beattie CE: Zebrafish deadly seven functions in neurogenesis. Dev Biol. 2001, 237: 306-323. 10.1006/dbio.2001.0381.
Hu ZL, Shi M, Huang Y, Zheng MH, Pei Z, Chen JY, Han H, Ding YQ: The role of the transcription factor Rbpj in the development of dorsal root ganglia. Neural Dev. 2011, 6: 14-10.1186/1749-8104-6-14.
Mead TJ, Yutzey KE: Notch pathway regulation of neural crest cell development in vivo. Dev Dyn. 2012, 241: 376-389. 10.1002/dvdy.23717.
Glavic A, Silva F, Aybar MJ, Bastidas F, Mayor R: Interplay between Notch signaling and the homeoprotein Xiro1 is required for neural crest induction in Xenopus embryos. Development. 2004, 131: 347-359.
High FA, Zhang M, Proweller A, Tu L, Parmacek MS, Pear WS, Epstein JA: An essential role for Notch in neural crest during cardiovascular development and smooth muscle differentiation. J Clin Invest. 2007, 117: 353-363. 10.1172/JCI30070.
Kubu CJ, Orimoto K, Morrison SJ, Weinmaster G, Anderson DJ, Verdi JM: Developmental changes in Notch1 and numb expression mediated by local cell-cell interactions underlie progressively increasing delta sensitivity in neural crest stem cells. Dev Biol. 2002, 244: 199-214. 10.1006/dbio.2002.0568.
Morrison SJ, Perez SE, Qiao Z, Verdi JM, Hicks C, Weinmaster G, Anderson DJ: Transient Notch activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest stem cells. Cell. 2000, 101: 499-510. 10.1016/S0092-8674(00)80860-0.
Taylor MK, Yeager K, Morrison SJ: Physiological Notch signaling promotes gliogenesis in the developing peripheral and central nervous systems. Development. 2007, 134: 2435-2447. 10.1242/dev.005520.
Woodhoo A, Alonso MB, Droggiti A, Turmaine M, D’Antonio M, Parkinson DB, Wilton DK, Al-Shawi R, Simons P, Shen J, Guillemot F, Radtke F, Meijer D, Feltri ML, Wrabetz L, Mirsky R, Jessen KR: Notch controls embryonic Schwann cell differentiation, postnatal myelination and adult plasticity. Nat Neurosci. 2009, 12: 839-847. 10.1038/nn.2323.
Tsarovina K, Schellenberger J, Schneider C, Rohrer H: Progenitor cell maintenance and neurogenesis in sympathetic ganglia involves Notch signaling. Mol Cell Neurosci. 2008, 37: 20-31. 10.1016/j.mcn.2007.08.010.
Okamura Y, Saga Y: Notch signaling is required for the maintenance of enteric neural crest progenitors. Development. 2008, 135: 3555-3565. 10.1242/dev.022319.
Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N: Differential Notch signaling distinguishes neural stem cells from intermediate progenitors. Nature. 2007, 449: 351-355. 10.1038/nature06090.
Shimojo H, Ohtsuka T, Kageyama R: Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron. 2008, 58: 52-64. 10.1016/j.neuron.2008.02.014.
Nikopoulos GN, Duarte M, Kubu CJ, Bellum S, Friesel R, Maciag T, Prudovsky I, Verdi JM: Soluble Jagged1 attenuates lateral inhibition, allowing for the clonal expansion of neural crest stem cells. Stem Cells. 2007, 25: 3133-3142. 10.1634/stemcells.2007-0327.
Adameyko I, Lallemend F, Aquino JB, Pereira JA, Topilko P, Müller T, Fritz N, Beljajeva A, Mochii M, Liste I, Usoskin D, Suter U, Birchmeier C, Ernfors P: Schwann cell precursors from nerve innervation are a cellular origin of melanocytes in skin. Cell. 2009, 139: 366-379. 10.1016/j.cell.2009.07.049.
Kruger GM, Mosher JT, Bixby S, Joseph N, Iwashita T, Morrison SJ: Neural crest stem cells persist in the adult gut but undergo changes in self-renewal, neuronal subtype potential, and factor responsiveness. Neuron. 2002, 35: 657-669. 10.1016/S0896-6273(02)00827-9.
Liu MT, Kuan YH, Wang J, Hen R, Gershon MD: 5-HT4 receptor-mediated neuroprotection and neurogenesis in the enteric nervous system of adult mice. J Neurosci. 2009, 29: 9683-9699. 10.1523/JNEUROSCI.1145-09.2009.
Nagoshi N, Shibata S, Kubota Y, Nakamura M, Nagai Y, Satoh E, Morikawa S, Okada Y, Mabuchi Y, Katoh H, Okada S, Fukuda K, Suda T, Matsuzaki Y, Toyama Y, Okano H: Ontogeny and multipotency of neural crest-derived stem cells in mouse bone marrow, dorsal root ganglia, and whisker pad. Cell Stem Cell. 2008, 2: 392-403. 10.1016/j.stem.2008.03.005.
Sieber-Blum M, Grim M, Hu YF, Szeder V: Pluripotent neural crest stem cells in the adult hair follicle. Dev Dyn. 2004, 231: 258-269. 10.1002/dvdy.20129.
Tomita Y, Matsumura K, Wakamatsu Y, Matsuzaki Y, Shibuya I, Kawaguchi H, Ieda M, Kanakubo S, Shimazaki T, Ogawa S, Osumi N, Okano H, Fukuda K: Cardiac neural crest cells contribute to the dormant multipotent stem cell in the mammalian heart. J Cell Biol. 2005, 170: 1135-1146. 10.1083/jcb.200504061.
Wong CE, Paratore C, Dours-Zimmermann MT, Rochat A, Pietri T, Suter U, Zimmermann DR, Dufour S, Thiery JP, Meijer D, Beermann F, Barrandon Y, Sommer L: Neural crest-derived cells with stem cell features can be traced back to multiple lineages in the adult skin. J Cell Biol. 2006, 175: 1005-1015. 10.1083/jcb.200606062.
Duff RS, Langtimm CJ, Richardson MK, Sieber-Blum M: In vitro clonal analysis of progenitor cell patterns in dorsal root and sympathetic ganglia of the quail embryo. Dev Biol. 1991, 147: 451-459. 10.1016/0012-1606(91)90303-K.
Hagedorn L, Suter U, Sommer L: P0 and PMP22 mark a multipotent neural crest-derived cell type that displays community effects in response to TGF-beta family factors. Development. 1999, 126: 3781-3794.
Le Lievre CS, Schweizer GG, Ziller CM, Le Douarin NM: Restrictions of developmental capabilities in neural crest cell derivatives as tested by in vivo transplantation experiments. Dev Biol. 1980, 77: 362-378. 10.1016/0012-1606(80)90481-9.
Li HY, Say EH, Zhou XF: Isolation and characterization of neural crest progenitors from adult dorsal root ganglia. Stem Cells. 2007, 25: 2053-2065. 10.1634/stemcells.2007-0080.
Rohrer H, Henke-Fahle S, el-Sharkawy T, Lux HD, Thoenen H: Progenitor cells from embryonic chick dorsal root ganglia differentiate in vitro to neurons: biochemical and electrophysiological evidence. EMBO J. 1985, 4: 1709-1714.
Singh RP, Cheng YH, Nelson P, Zhou FC: Retentive multipotency of adult dorsal root ganglia stem cells. Cell Transplant. 2009, 18: 55-68. 10.3727/096368909788237177.
Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF: Stages of embryonic development of the zebrafish. Dev Dyn. 1995, 203: 253-310. 10.1002/aja.1002030302.
Obholzer N, Wolfson S, Trapani JG, Mo W, Nechiporuk A, Busch-Nentwich E, Seiler C, Sidi S, Söllner C, Duncan RN, Boehland A, Nicolson T: Vesicular glutamate transporter 3 is required for synaptic transmission in zebrafish hair cells. J Neurosci. 2008, 28: 2110-2118. 10.1523/JNEUROSCI.5230-07.2008.
Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB: The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007, 236: 3088-3099. 10.1002/dvdy.21343.
Kikuta H, Kawakami K: Transient and stable transgenesis using tol2 transposon vectors. Methods Mol Biol. 2009, 546: 69-84. 10.1007/978-1-60327-977-2_5.
Ungos JM, Karlstrom RO, Raible DW: Hedgehog signaling is directly required for the development of zebrafish dorsal root ganglia neurons. Development. 2003, 130: 5351-5362. 10.1242/dev.00722.
Marusich MF, Furneaux HM, Henion PD, Weston JA: Hu neuronal proteins are expressed in proliferating neurogenic cells. J Neurobiol. 1994, 25: 143-155. 10.1002/neu.480250206.
Brend T, Holley SA: Zebrafish whole mount high-resolution double fluorescent in situ hybridization. J Vis Exp. 2009, pii: 1229-
Bierkamp C, Campos-Ortega JA: A zebrafish homologue of the Drosophila neurogenic gene Notch and its pattern of transcription during early embryogenesis. Mech Dev. 1993, 43: 87-100. 10.1016/0925-4773(93)90027-U.
Appel B, Eisen JS: Regulation of neuronal specification in the zebrafish spinal cord by Delta function. Development. 1998, 125: 371-380.
Haddon C, Smithers L, Schneider-Maunoury S, Coche T, Henrique D, Lewis J: Multiple delta genes and lateral inhibition in zebrafish primary neurogenesis. Development. 1998, 125: 359-370.
Ma EY, Rubel EW, Raible DW: Notch signaling regulates the extent of hair cell regeneration in the zebrafish lateral line. J Neurosci. 2008, 28: 2261-2273. 10.1523/JNEUROSCI.4372-07.2008.
Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, Bazett-Jones DP, Allis CD: Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997, 106: 348-360. 10.1007/s004120050256.
Laguerre L, Soubiran F, Ghysen A, Konig N, Dambly-Chaudiere C: Cell proliferation in the developing lateral line system of zebrafish embryos. Dev Dyn. 2005, 233: 466-472. 10.1002/dvdy.20343.
Poss PD, Wilson LG, Keating MT: Heart regeneration in the zebrafish. Science. 2002, 298: 2188-2190. 10.1126/science.1077857.
Harris JA, Cheng AG, Cunningham LL, MacDonald G, Raible DW, Rubel EW: Neomycin-induced hair cell death and rapid regeneration in the lateral line of zebrafish (Danio rerio). J Assoc Res Otolaryngol. 2003, 4: 219-234. 10.1007/s10162-002-3022-x.
Acknowledgements
We thank Bruce Appel for the Tg(hsp70:XdnSu(H)myc) line and for the sox10 promoter, Teresa Nicolson for the p5E-neuroD construct, and Chi-Bin Chien for the pME-TagRFP construct. We thank David White and fish facility staff for excellent fish care. Work was supported by National Institutes of Health grants R01 NS057220 (DWR), T32 GM007270 (AP) and T32 HD007183 (HFM).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
HFM, CDS and DWR designed and analyzed experiments. HFM, AP and CDS performed experiments. AP, AS and AN developed key reagents. HFM, CDS and DWR wrote the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
Additional file 1: Nascent DRG neurons arise by differentiation. Time-lapse confocal projections showing sensory neuron differentiation in a TgBAC(neurod:EGFP) embryo between 44 and 52 hpf. The embryo was imaged every six minutes for eight hours. TgBAC(neurod:EGFP) is expressed in newly formed DRG neurons. At time zero, there are one to two GFP + cells per ganglion. During the course of imaging, GFP + cells are added to the ganglia by upregulation of the transgene. No GFP + cells are seen to divide during imaging. Scale bar, 20 μm. (MOV 413 KB)
13064_2012_204_MOESM2_ESM.mov
Additional file 2: Migrating neural crest cells form the DRG anlage. Time-lapse confocal projections of a wild-type embryo expressing Tg(sox10:nlsEos) transgene in migrating neural crest cells. The embryo was imaged every 10 minutes for 23 hours beginning at 18 hpf. During this time, a subset neural crest cells migrate into the region of the forming DRG and remain there. No neural crest cells migrated into this region after 10 hours of imaging at approximately 28 hpf. Scale bar, 25 μm. (MOV 5 MB)
13064_2012_204_MOESM3_ESM.mov
Additional file 3: Neural crest cells divide and differentiate as neurons in the DRG. Time-lapse confocal projections of the forming DRG in a Tg(sox10:nlsEos)/Tg(neurod:TagRFP) transgenic embryo between 30 and 56.5 hpf. The embryo was imaged every 10 minutes for 26:30 hours. One cell upregulates Tg(neurod:TagRFP) as it differentiates as a neuron. Area boxed at 26:30 is shown again as a 4D projection in close-up, then in reverse with the neuronal precursor tracked from the progenitor cell division. (MOV 11 MB)
13064_2012_204_MOESM4_ESM.mov
Additional file 4: Proliferating cells are associated with late larval DRG. Confocal 1.5 μm slices through an individual ganglion in a 25 dpf, 6.25 mm larval fish. Cells that have incorporated BrdU (red) or are labeled with anti-pH3 antibody (blue) intermingle with Elavl labeled neurons (green). Neurons are not co-labeled with markers of proliferation. Scale bar, 20 μm. (MOV 357 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
McGraw, H.F., Snelson, C.D., Prendergast, A. et al. Postembryonic neuronal addition in Zebrafish dorsal root ganglia is regulated by Notch signaling. Neural Dev 7, 23 (2012). https://doi.org/10.1186/1749-8104-7-23
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1749-8104-7-23