Abstract
Background
Widespread in the environment, Staphylococcus spp. infect animals and humans as normal flora or pathogens. By extending our recent report of multi-drug resistant (MDR) S. aureus in dairy goats, this study investigated the staphylococcal infection and characterized the MDR-S. aureus and methicillin-resistant S. aureus (MRSA) isolates collected from goats in 2008 to elucidate the appearance of MRSA in goats and the mastitis associated staphylococcus enterotoxin (SE) types. A total of 555 samples were collected from six goat parts and three environmental sources among four dairy goat farms in southern Taiwan. Coagulase-positive and negative Staphylococcus spp. (CPS and CNS, respectively) were also identified. Furthermore, predominant SE genes of nine enterotoxin genes sea through sej along with antimicrobial resistance and genetic variations were determined.
Results
In total, 137 staphylococcal strains were identified and found predominantly in milk, and in the vagina, anus, and nasal cavity. The most prevalent species was S. lentus, followed by S. aureus, S. epidermidis, and S. xylosus. Enterotoxin genes were not identified in any CNS isolates, however sec and see were identified only in S. aureus associated with mastitis in goat. In compared to the isolates from 2006 to 2007, 27 S. aureus isolates from 2008 were found to be more resistant to ampicillin, cephalothin, oxacillin, oxytetracycline, penicillin G, and tetracycline. Eleven MRSA isolates were identified and belonged to SCCmec type III (nine isolates) as the major type and SCCmec type II (two isolates). These MRSA isolates revealed pulse-field gel electrophoresis (PFGE) pattern A (five isolates), C (one isolate), and D (one isolate) of human isolates. The other two isolates without pulsotypes belonged to ST59.
Conclusion
The prevalence and infection sites of CNS differed from those of CPS. Genetic analyses indicated that genetic divergence, possible zoonotic transfer of MRSA, and the involvement of sec as important virulence factors for of S. aureus that lead to mastitis in goats.
Similar content being viewed by others
Background
Important pathogenic species of coagulase-positive and negative Staphylococcus (CPS and CNS, respectively) are S. aureus and S. epidermidis, respectively, which are frequently isolated from the environment and infected animals and humans [1, 2]. S. aureus can cause severe blood infections and necrotizing fasciitis in humans, wound infection and mastitis in cattle, horses, pigs, and goats, exudative epidermitis in pigs, pyoderma in horses, dogs, and cats, pyemic sheep [2, 3]. As an opportunistic pathogen, CNS occasionally cause subclinical or clinical mastitis. For example, in cattle mastitis can be caused by S. capitis, S. chromogenes, S. cohnii, S. epidermidis, S. haemolyticus, S. hominis, S. simulans, S. warneri, S. hyicus, and S. caseolyticus; in goats by S. caprae; and in cattle and sheep by S. xylosus [2]. However, CNS can damage breast tissue to increase the somatic cell counts and decrease milk quality and production [4]. The identification of CNS species is essential to determine their pathogenicity and to develop management practices to prevent mastitis. Due to difficult and expensive procedures for identifying these organisms, many laboratories do not perform these assays.
As opportunistic pathogens, S. aureus and S. epidermidis can also cause bacteremia, subacute endocarditis, and can form biofilms on heart valve prostheses, shunts, total joint arthroplasty or surgical sutures in humans [5]. Some S. aureus isolates possess a staphylococcal enterotoxin (SE), which is a virulence factor for foodborne disease and can form several products from SE genes, designates sea - seu [6, 7]. These SE genes are associated with host-specific infection, such as seb for foodborne infection in humans, sea for bovine infection [8], and sec for mastitis in goats and bovine [9]. However, more than 10 SE genes were identified in isolates obtained from goats [7]. As the most prevalent pathogen in both hospitals and communities, S. aureus can become methicillin (oxacillin)-resistant S. aureus (MRSA/ORSA) by the introduction of an exogenous mobile staphylococcal chromosomal cassette, mec (SCCmec), encoding a low-affinity penicillin-binding protein 2a responsible for methicillin/oxacillin resistance [10–14]. Based on variations in the ccr operon and mec complexes, at least seven SCCmec types have been identified [14–16]. Moreover, methicillin-resistant CNS has been identified in animals [17].
The extensive use of antibiotics in hospitals and animals has increased the emergence of MDR Staphylococcus [1]. Since the emergence of MRSA isolated from humans in the 1960s [18], MRSA has been frequently isolated in hospitals, dogs, and horses as a zoonotic pathogen [17, 19, 20]. Our recent study characterized MDR S. aureus from dairy goats in 2006-2007 and no MRSA was isolated [21]. The current study investigated the prevalent Staphylococcus spp. and characterized the MDR-S. aureus and MRSA isolates collected from goats in 2008 by nucleotide sequencing, pulse-field gel electrophoresis (PFGE) analysis and polymerase chain reaction (PCR) analysis to elucidate the origins of MRSA isolates and the SE types associated with mastitis in goats.
Results
Identification of staphylococcusspecies
In total, 555 samples were collected from six bodily regions of dairy goats and three environmental sources in 2008. Among 45.4% (252/555) samples with bacterial growth on blood agar, 137 staphylococcal strains and 115 non-staphylococcal strains were identified. Five major Staphylococcus spp. were identified and listed in decreasing order of isolate number: S. lentus (31), S. aureus (27), S. epidermidis (23), S. xylosus (17) and S. caprae (3), and others (36).
Prevalent staphylococcusspecies in different sampling locations and farms
Staphylococcal species were frequently isolated from milk, anus, vagina, nasal cavity, and udders (Table 1). However, the predominant infection sites differed between S. aureus and CNS. On average, the prevalence of S. aureus was 18.9 and 0.7% for goats and environmental samples, respectively, and differed among farms and body regions of goats (Table 1). In contrast to being absent in the anus and dorsum, S. aureus was predominant in the nasal cavity (12), vagina (9), and milk (4); in addition, CNS was prevalent in the milk (54), vagina (26), and anus (26).The highest prevalence of S. aureus was found in Farm A, followed by Farms D, C and B. In contrast, the highest prevalence of CNS was found in goats on Farm D followed by Farms C, B, and A.
Antimicrobial susceptibility
Fifteen antimicrobial agents were used to characterize S. aureus strains and could be divided into 13 antibiograms. Nearly all isolates were susceptible to enrofloxacin, gentamycin, neomycin, and vancomycin; in addition, over 60% of the isolates were also resistant to penicillin (P), ampicillin (A), cephalothin, tetracycline (T), and oxytetracycline (Ot) (Table 2). In this study, 11 (40.7%) MRSA isolates were identified in nasal cavity, vagina, and goat milk only. Isolates from the nasal cavity exhibited the highest antimicrobial resistance. Compared with the antimicrobial resistance found for isolates identified in 2006-2007, isolates from 2008 revealed an increase in resistance to ampicillin, bacitracin (B), cloxacin (Cl), oxacillin (Ox), penicillin G, and streptomycin (S) (Table 2). Although 88.9% of isolates were resistant to more than one antimicrobial agent; the most prevalent antibiograms were AOtPT (29.6%, 8/27) and ABClOtOxPST (25.9%, 7/27). The antibiogram number varied among farms from six in Farm A to five in Farm B, four in Farm D and two in Farm C. Differences in antibiograms and their number may indicate the different bacterial sources in each farm. The minimum inhibitory concentrations (MIC) of 11 MRSA isolates to methicillin ranged from 16 to 256 μg/mL, and varied among farms and bodily region. The highest MIC was found in isolates collected from the nasal cavity (64-256 μg/mL), followed by goat milk (64 μg/mL), and vagina (16-64 μg/mL).
Staphylococcal enterotoxin types
Of the nine enterotoxin genes examined, only sec and see were identified in S. aureus isolates and no SE genes were found in 71 CNS strains (Table 3). Eight out of 10 S. aureus strains isolated from milk of 15 goats with mastitis were shown to possess sec. PCR-restriction fragment length polymorphism (RFLP) analysis of sec PCR products revealed an identical AluI restriction pattern for all strains. In 27 S. aureus bovine strains, only sea was identified in three strains.
Characterization of oxacillin-resistant S. aureus
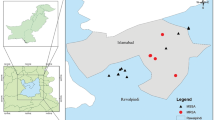
PCR amplification of 11 MRSA isolates identified SCCmec type III in nine isolates from Farms A (8) and B (1), and SCCmec type II in two isolates found in Farms B (1) and D (1) (Table 4). Two isolates could not be pulsotyped and six pulsotypes were determined (Figure 1). Four pulsotypes were identified in six isolates of Farm A, revealing identical patterns as pulsotypes A, C, and D in human isolates previously identified in Taiwan [22]. The remaining isolates belonged to pulsotypes VIA and VIB (no similar pulsotypes as human isolates) in Farm B and pulsotype V (pulsotype A of human isolate) in Farm D. Multi-locus sequence typing (MLST) analysis indicated that two isolates for which no pulsotype could be determined were ST59, the common MLST types of human isolates. Phylogenetic analysis of sec showed that sequence similarity ranged from 94.8-100% (Figure 2).
Discussion
CNS strains S. lentus, S. epidermidis, S. xylosus, and S. caprae, and S. aureus were identified as the predominant species infecting goats in this study. S. aureus and CNS differed in prevalence and bodily location in goats (Table 1). Additionally, CNS and S. aureus accounted for 38.2 and 11% of infections, respectively, in the milk of goats with mastitis [23], and 2.6% (35/1,372) and 17.3% (237/1,372), respectively, in the milk of dairy cows and goats in Taiwan [24]. In animals, the prevalence of S. aureus differed among countries, such as 2.2% in the United Kingdom [25], 6.7% in Canada [26], 9.1% in USA [27], 24.2% in Ethiopia [28], 29.6% in Korea [29], and 32.9% in Uruguay [30]. However, S. aureus accounts for 18.7% (64/342) of staphylococcal infections in goats [7]. In Taiwan, S. aureus infection in goats increased from 1.7% (12/706) [24] up to 2.5% (86/3,427) [31] and 4.9% (27/555) in this study. Sanitation may be another important factor that leads to an increase in S. aureus infection. Indeed, sanitation could reduce the S. aureus infection from 6.6 to 0.6% or from 14.6 to 3.0% [32, 33]. In Taiwan, S. aureus infection in milk ranged from 0-5.2% among farms [21]. In the present study, the prevalence ranged from 3.1 to 6.8% for S. aureus and from 11.2 to 31.8% for CNS among farms, suggesting that CNS infection is more common in goats.
Early reports indicated that staphylococcal infections in goat bodily regions differed and ranged from 6.1% (21/342) in the armpits up to 70.8% (242/342) on the skin of udders and mamilla, and contributed to only 0.7% contamination of the apparatus [7]. Although lacking statistical analysis in this study, Staphylococcus was identified in 42.3% (58/137) and 5.8% (8/137) of milk and udders, respectively (Table 1), suggesting that Staphylococcus is a major bacterial cause of mastitis in goats that can then infect humans through unsanitary milk. As an important virulence factor causing foodborne disease in humans, predominant SE genes are associated with outbreaks in certain countries, such as sea in France [34], seb in eastern Slovakia, Tehran, and Japan [35–37], sec-2 and sec-3 in Taiwan [38]. Furthermore, particular sec types is also associated with food-related Staphylococcus spp. [39], in S. aureus, and CNS isolated from the milk of sheep [7, 40], and in S. aureus collected from sheep or cows with mastitis [41]. Additionally, sea was determined to be the major type in S. aureus from cows with mastitis [8]. As shown in Table 3, sec and sea were the major SE genotypes found in goats and cows, respectively, and sec was associated with mastitis in goats. These data confirmed the importance of the sec gene involved in goat mastitis pathogenesis [42]. In cattle, the SEC toxin (not toxic shock syndrome toxin type 1 or TSST-1) was previously shown to significantly increase somatic cell counts and enhance the severity of the mastitis in acute mastitis [43]. Although SE genes have been found in CNS species, such as S. xylosus, S. warneri, and S. chromogenes isolated from cows and goats [7, 40, 44, 45], they were only identified in CPS but not CNS isolated previously in Taiwan [46, 47] and in the present study (Table 3). As an opportunistic pathogen that can cause mastitis in cattle and goats [1, 21, 48, 49], S. aureus infection can also cause clinical symptoms in cattle [50] but is typically asymptomatic in goats [21, 48].
Using penicillin, ampicillin, and tetracycline antibiotics to treat bacterial infections in animals often increases resistance to these antibiotics. In cattle, S. aureus or other Staphylococcus spp. causing mastitis were more resistance to penicillin and ampicillin, streptomycin, tetracycline, and oxytetracycline [1, 24, 49, 51]. Our data showed an increased resistance to penicillin, ampicillin, cloxacillin, and cephalothin from 2006 to 2008, in addition to the appearance of MRSA isolates from goats in 2008 (Table 2). Furthermore, the MRSA isolates identified in this study belonged to the major nosocomial SCCmec types: SCCmec type II and III [52, 53]. Zoonotic transfer of MRSA has been reported between horses and humans in the USA [20, 54], between cattle and humans in Korea [55], and between livestock and humans in Taiwan [56, 57]. Our data indicate that MRSA isolates may have been acquired from humans or transmitted from different goat breeding farms.
PFGE analysis is typically performed to trace the pathogens responsible for outbreaks. Containing a thick cell wall, S. aureus must be treated with lysostaphin, not lysozyme, to break the pentaglycine linkage within the peptidoglycan [58, 59]. For genomic analysis of S. aureus, genomic DNA cannot be digested by restriction enzyme SmaI and diverse genomic variations in size [60] limit the utility of PFGE analysis. However, pulsotypes appear to correlate with human disease. Pulsotype D S. aureus is associated with more severe symptoms than pulsotype type C bacteria that only cause mild symptoms [50]. In Taiwan, MRSA accounts for 53-83% of S. aureus isolates from hospitals [61] and the major pulsotypes of human MRSA are pulsotype A, followed by types C and D [22]. In this study, pulsotype A was also the most prevalent type of goat MRSA isolate (Figure 1, Table 4). Additionally, PFGE analysis also revealed diverse sources of MRSA in Farm A and a single origin in Farms B and D (Table 4).
Conclusion
The current study was the first report of the appearance of MRSA strains and sec-associated mastitis in goats from Taiwan. Analysis of SCCmec types and pulsotypes revealed that the genetically diverse MRSA strains might have been acquired from humans or transferred from different goat breeding farms.
Methods
Source of bacterial isolates
Samples were acquired aseptically according to the procedure described by the National Mastitis Council (NMC) with some modification [62]. Informed consent was obtained from all farm owners prior to the start of this study. A total of 555 samples were collected from the milk, anus, dorsum, nasal cavity, udders, and vagina of goats, in addition to milking apparatus, bulk tank, and water of environments at four different goat farms in Taiwan during 2008. Isolates (n = 137) were characterized by culture on blood agar [63], morphology, Gram stain, catalase test, and growth on Staphylococcus medium No. 110 (Oxoid, Basingstoke, UK). Furthermore, Staphylococcus species were biotyped according to method of Myllys et al. [64] using the API Staph identification kit (BioMerieux, Marcy l'Etoile, France). Additionally, 86 isolates collected in 2006-2007 were compared with those isolated in 2008. CPS (BCRC 14958, ATCC 27664) and CNS (BCRC 15228) strains were purchased from the Bioresource Collection and Research Center (BCRC, Taiwan). Twenty-seven S. aureus strains isolated from cattle with mastitis were kindly provided by Professor Shih-Te Chuang, (College of Veterinary Medicine, National Chung Hsing University, Taiwan) for examination of enterotoxin genotypes. Human isolates were kindly provided by Professor Chishih Chu, (the Department of Microbiology, Immunology, and Biopharmaceuticals, National Chiayi University). The use of all bacteria was supervised by the Biological Security Committee of National Chiayi University in accordance with the laws of Taiwan. The protocols and regulations used during the study were performed according to the guidelines of"Animal Use Protocol, the Institutional Animal Care and Use Committee (IACUC), National Chiayi University" with great care for animal welfare and adherence to the law.
Antimicrobial susceptibility test
The disc diffusion method and the guidelines of the Clinical and Laboratory Standards Institute (CLSI) standards and the manufacturer were used to determine the susceptibility of each isolate to ampicillin (AMP; 10 μg), bacitracin (BAC; 10 units), oxacillin (OXA; 1 μg), cefuroxime (CXM; 30 μg), cephalothin (CEP; 30 μg), cloxacillin (CLO; 5 μg), enrofloxacin (ENR; 5 μg), gentamicin (GEN; 10 μg), neomycin (NEO; 30 μg), oxytetracycline (OXY; 30 μg), penicillin G (PEN; 10 U), streptomycin (STR; 10 μg), sulfamethoxazole/trimethoprim (Sxt; 23.75 μg for S and 1.25 μg for t), tetracycline (TET; 30 μg), and vancomycin (VAN; 30 μg) [65]. Results of the antimicrobial susceptibility were also validated using Escherichia coli (ATCC No. 25922). Bacto discs were purchased from Becton Dickinson (Sparks, MD, USA). Finally, MIC to oxacillin of each ORSA isolate was determined by Etest (AB Biodisk, Solna, Sweden).
SCCmectyping
DNA templates of MRSA isolates were prepared for PCR amplification using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA). Table 5 lists the primers used to identify mecA and the SCCmec types [52, 66, 67]. A 50-μL PCR reagent included 5 μL of DNA template, 20 μM of each primer, 0.2 mM dNTPs, 5 μL of 10× PCR reaction buffer, 1.4 U Taq DNA polymerase, and 33.3 μL distilled water. The PCR conditions were a initial denaturation at 94°C for 4 min, followed by 30 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 1 min and a final extension at 72°C for 4 min. PCR products were separated by 2% agarose at 50 V for 1 hr and visualized under ultraviolet illumination after ethidium bromide (EtBr) staining.
PCR identification of SE types and PCR-RFLP analysis
Staphylococcal enterotoxin genes, sea-sej, were amplified by PCR using primers listed in Table 5[68–76]. 50-μL PCR reagent was described earlier. The cycling conditions for multiplex PCR were 5 min at 94°C for initial denaturation, followed by 30 cycles of 1 min at 94°C, 30 s at 60°C, 1 min at 72°C and a final extension at 72°C for 10 min to amplify sea - see. For amplification of seg-sej, initial denaturation was performed for 3 min at 94°C, followed by 30 cycles of 30 s at 94°C, 30 s at 60°C, and 30 s at 72°C and a final extension at 72°C for 10 min. PCR products of sec were purified from agarose gels using the Gene-Spin-V2 Miniprep purification kit (Protech Technology, Solon, OH, USA), and the purified products were digested with restriction enzyme AluI and separated by 1% agarose gel run at 50 V for 1 hr. Furthermore, the purified PCR products were sequenced and analyzed using the Megalign program of the Lasergene software (DNAstar, Madison, Wisconsin, USA).
Genetic typing
The genotype of each MRSA isolate was determined by restriction enzyme SmaI-digested PFGE analysis, as previously described [77]. Briefly, whole-cell embedded agarose plugs were digested with SmaI and then resolved by a CHEF DR-III apparatus (Bio-Rad, Hercules, CA, USA). Finally, the MRSA isolate BCRC 15211 was used as the standard size marker. Pulsotypes and MLST types of MRSA were determined based on the method described by Enright et al. [78] and analysis of MLST databases, respectively.
Authors' information
Chishih Chu is a Professor in the Department of Microbiology, Immunology, and Biopharmaceuticals. Changyou Yu is a Professor in the Department of Veterinary Medicine. Yanhaui Lee was a Master student in the Department of Veterinary Medicine. Yaochi Su is an Associate Professor in the Department of Veterinary Medicine, National Chiayi University, Taiwan, Republic of China.
Abbreviations
- AMP:
-
ampicillin
- BAC:
-
bacitracin
- CEP:
-
cephalothin
- CLO:
-
cloxacillin
- CXM:
-
cefuroxime
- ENR:
-
enrofloxacin
- EtBr:
-
ethidium bromide
- GEN:
-
gentamicin
- MDR:
-
multi-drug resistance
- MIC:
-
minimum inhibitory concentration
- OXA:
-
oxacillin
- OXY:
-
oxytetracycline
- PCR-RFLP:
-
polymerase chain reaction- restriction fragment length polymorphism
- PEN:
-
penicillin G
- PFGE:
-
pulsed-field gel electrophoresis
- STR:
-
streptomycin
- Sxt:
-
sulfamethoxazole/trimethoprim
- TET:
-
tetracycline
- VAN:
-
vancomycin.
References
Resch M, Nagel V, Hertel C: Antibiotic resistance of coagulase-negative staphylococci associated with food and used in starter cultures. Int J Food Microbiol. 2008, 127: 99-104. 10.1016/j.ijfoodmicro.2008.06.013.
Quinn PJ, Markey BK, Carter ME, Donnelly WJ, Leonard FC, Maguire D: Staphylococcus species. Veterinary Microbiology and Microbial Disease. Edited by: Quinn PJ. 2002, USA: Blackwell Science Ltd, 43-48. 1
Abigail AS, Whitt DD: Bacterial Pathogenesis: A Molecular Approach. 2001, Washington: ASM Press
Taponen S, Pyorala S: Coagulase-negative staphylococci as cause of bovine mastitis-not so different from Staphylococcus aureus?. Vet Microbiol. 2009, 134: 29-36. 10.1016/j.vetmic.2008.09.011.
Krimmer V, Merkert H, von Eiff C, Frosch M, Eulert J, Löhr JF, Hacker J, Ziebuhr W: Detection of Staphylococcus aureus and Staphylococcus epidermidi in clinical samples by 16S rRNA-directed in situ hybridization. J Clin Microbiol. 1999, 37: 2667-2673.
Løvseth A, Loncarevic S, Berdal KG: Modified multiplex PCR method for detection of pyrogenic exotoxin genes in staphylococcal isolates. J Clin Microbiol. 2004, 42: 3869-3872. 10.1128/JCM.42.8.3869-3872.2004.
Valle J, Gomez-Lucia E, Piriz S, Goyache J, Orden JA, Vadillo S: Enterotoxin production by Staphylococci isolated from healthy goats. Appl Environ Microbiol. 1990, 56: 1323-1326.
Lim SK, Joo YS, Moon JS, Lee AR, Nam HM, Wee SH, Koh HB: Molecular typing of enterotoxigenic Staphylococcus aureus isolated from bovine mastitis in Korea. J Vet Med Sci. 2004, 66: 581-584. 10.1292/jvms.66.581.
da Silva ER, do Carmo LS, da Silva N: Detection of the enterotoxins A, B, and C genes in Staphylococcus aureus from goat and bovine mastitis in Brazilian dairy herds. Vet Microbiol. 2005, 106: 103-107. 10.1016/j.vetmic.2004.12.005.
Reynolds PE, Brown DF: Penicillin-binding proteins of [beta]- lactam- resistant strains of Staphylococcus aureus: effect of growth conditions. FEBS Lett. 1985, 192: 28-32. 10.1016/0014-5793(85)80036-3.
Geha DJ, Uhl JR, Gustaferro CA, Persing DH: Multiplex PCR for identification of methicillin-resistant staphylococci in the clinical laboratory. J Clin Microbiol. 1994, 32: 1768-1772.
Katayama Y, Ito T, Hiramatsu K: A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000, 44: 1549-1555. 10.1128/AAC.44.6.1549-1555.2000.
Oliveira DC, de Lencastre H: Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002, 46: 2155-2161. 10.1128/AAC.46.7.2155-2161.2002.
Chongtrakool P, Ito T, Ma XX, Kondo Y, Trakulsomboon S, Tiensasitorn C, Jamklang M, Chavalit T, Song JH, Hiramatsu K: Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob Agents Chemother. 2006, 50: 1001-1012. 10.1128/AAC.50.3.1001-1012.2006.
Boyle-Vavra S, Yin S, Challapalli M, Daum RS: Transcriptional induction of the penicillin-binding protein 2 gene in Staphylococcus aureus by cell wall-active antibiotics oxacillin and vancomycin. Antimicrob Agents Chemother. 2003, 47: 1028-1036. 10.1128/AAC.47.3.1028-1036.2003.
Oliveira DC, Milheirico C, de Lencastre H: Redefining a structural variant of staphylococcal cassette chromosome mec, SCCmec type VI. Antimicrob Agents Chemother. 2006, 50: 3457-3459. 10.1128/AAC.00629-06.
van Duijkeren E, Box AT, Heck ME, Wannet WJ, Fluit AC: Methicillin-resistant Staphylococci isolated from animals. Vet Microbiol. 2004, 103: 91-97. 10.1016/j.vetmic.2004.07.014.
Barrett FF, McGehee RF, Finland M: Methicillin-resistant Staphylococcus aureus at Boston City Hospital - bacteriologic and epidemiologic observations. N Engl J Med. 1968, 279: 441-448. 10.1056/NEJM196808292790901.
Lin YC: Analysis of molecular typing Staphylococcus aureus of isolated from bacterimia patients. Master Thesis. 2005, Tzu Chi University, Graduate Institute of Medical Science
Seguin JC, Walker RD, Caron JP, Kloos WE, George CG, Hollis RJ, Jones RN, Pfaller MA: Methicillin-resistant Staphylococcus aureus outbreak in a veterinary teaching hospital: potential human-to-animal transmission. J Clin Microbiol. 1999, 37: 1459-1463.
Chu C, Lai JM, Yu CY, Chang WS, Lin HH, Su YC: Rapid identification of staphylococcal infection in dairy goat raw milk by a multiplex PCR assay. Taiwan Vet J. 2010, 36: 182-190.
Huang YC, Su LH, Wu TL, Liu CE, Young TG, Chen PY, Hseuh PR, Lin TY: Molecular epidemiology of clinical isolates of methicillin-resistant Staphylococcus aureus in Taiwan. J Clin Microbiol. 2004, 42: 307-310. 10.1128/JCM.42.1.307-310.2004.
White EC, Hinckley LS: Prevalence of mastitis pathogens in goat milk. Small Ruminant Res. 1999, 33: 117-121. 10.1016/S0921-4488(99)00013-9.
Tung CC: Application of PCR-RFLP on molecular epidemiological study of Staphylococcus aureus isolated from milk of dairy cows and goats. Master thesis. 2004, National Chung Hsing University, Department of Veterinary Medicine
Bradley AJ, Green MJ: A study of the incidence and significance of Gram-positive infections acquired during the dry period under UK field conditions. Proceedings of the 40th Annual Meeting of the National Mastitis Council: 11-14 February 2001. 2001, Nevada: Reno, 185-186.
Sargeant JM, Scott HM, Leslie KE, Ireland MJ, Bashiri A: Clinical mastitis in dairy cattle in Ontario: frequency of occurrence and bacteriological isolates. Can Vet J. 1998, 39: 33-38.
Wilson DJ, Gonzalez RN, Das HH: Bovine mastitis pathogens in New York and Pennsylvania: prevalence and effects on somatic cell count and milk production. J Dairy Sci. 1997, 80: 2592-2598. 10.3168/jds.S0022-0302(97)76215-5.
Workineh S, Bayleyegn M, Mekonnen H, Potgieter LN: Prevalence and aetiology of mastitis in cows from two major Ethiopian dairies. Trop Anim Health Prod. 2002, 34: 19-25. 10.1023/A:1013729626377.
Lee JH: Methicillin (Oxacillin)-resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl Environ Microbiol. 2003, 69: 6489-6494. 10.1128/AEM.69.11.6489-6494.2003.
Gianneechini R, Concha C, Rivero R, Delucci I, Moreno LJ: Occurrence of clinical and sub-clinical mastitis in dairy herds in the west littoral region in Uruguay. Acta Vet. 2002, 43: 221-230. 10.1186/1751-0147-43-221.
Lin HH: Epidemiology for rapid identification, and genotyping, and antibiotic susceptibility of Staphylococcus aureus isolated from raw milk in dairy goats. Master thesis. 2008, National Chiayi University, Department of Veterinary Medicine
Erskine RJ, Eberhart RJ, Hutchinson LJ, Spencer SB: Herd management and prevalence of mastitis in dairy herds with high and low somatic cell counts. J Am Vet Med Assoc. 1987, 190: 1411-1416.
Hutton CT, Fox LK, Hancock DD: Mastitis control practices: differences between herds with high and low milk somatic cell counts. J Dairy Sci. 1990, 73: 1135-1143. 10.3168/jds.S0022-0302(90)78774-7.
Kérouanton A, Hennekinne JA, Letertre C, Chesneau O, Brisabois A, De Buyser ML: Characterization of Staphlococcus aureus strains associated with food poisoning outbreaks in France. Int J Food Microbiol. 2007, 115 (3): 369-375. 10.1016/j.ijfoodmicro.2006.10.050.
Holecková B, Holoda E, Fotta M, Kalinácova V, Gondoĺ J, Grolmus J: Occurrence of enterotoxigenic Staphlococcus aureus in food. Ann Agri Environ Med. 2002, 9 (2): 179-182.
Pourmand MR, Memariani M, Hoseini M, Yazdchi SB: High prevalence of SEA gene among clinical isolates of Staphlococcus aureus in Tehran. Acta Med Iran. 2009, 47 (5): 357-361.
Mizumachi E, Kato F, Hisatsune J, Tsuruda K, Uehara Y, Seo H, Sugai M: Clonal distribution of enterotoxingenic Staphlococcus aureus on handles of handheld shopping baskets in supermarkets. J Appl Microbial. 2011, 110 (2): 562-567. 10.1111/j.1365-2672.2010.04910.x.
Chen TR, Hsiao MH, Chiou CS, Tsen HY: Development and use of PCR primers for the investigation of C1, C2 and C3 enterotoxin types of Staphylococcus aureus strains isolated from food-borne outbreaks. Int J Food Microbiol. 2001, 71: 63-70. 10.1016/S0168-1605(01)00564-5.
Rosec JP, Guiraud JP, Dalet C, Richard N: Enterotoxin production by Staphylococci isolated from foods in France. Int J Food Microbiol. 1997, 35: 213-221. 10.1016/S0168-1605(96)01234-2.
Bautista L, Gaya P, Medina M, Nenez M: A quantitative study of enterotoxin production by sheep milk Staphylococci. Appl Environ Microbiol. 1988, 54: 564-569.
Orden JA, Goyache J, Hernandez J, Domenech A, Suarez G, Gomez LE: Detection of enterotoxins and TSST-1 secreted by Staphylococcus aureus isolated from ruminant mastitis, comparison of ELISA and immunoblot. J Appl Bacteriol. 1992, 72: 486-489. 10.1111/j.1365-2672.1992.tb01863.x.
da Silva ER, do Carmo LS, da Silva N: Detection of the enterotoxins A, B, and C genes in Staphylococcus aureus from goat and bovine mastitis in Brazilian dairy herds. Vet Microbiol. 2005, 106: 103-107. 10.1016/j.vetmic.2004.12.005.
Kuroishi T, Komine KL, Kai K, Itagaki M, Koboyashi J, Ohta M, Kamata SK, Kumagai K: Concentrations and specific antibodies to staphylococcal enterotoxin-C and toxic shock syndrome toxin-1 in bovine mammary gland secretions, and inflammatory response to the intramammary inoculation of these toxins. Journal of Vet Med Sci. 2003, 65: 899-906. 10.1292/jvms.65.899.
Tondo EC, Guimaraes MC, Henriques JA, Ayub MA: Assessing and analyzing contamination of a dairy products processing plant by Staphylococcus aureus using antibiotic resistance and PFGE. Can J Microbiol. 2000, 46: 1108-1114.
White DG, Harman RJ, Matos JES, Langlois BE: Isolation and identification of coagulase negative staphylococcus species from bovine body sites and streak canals of nulliparous heifers. J Dairy Sci. 1989, 72: 1886-1892. 10.3168/jds.S0022-0302(89)79307-3.
Wang SJ, Chow LW, Chen JH, Liang I: Distribution and identification of enterotoxigenic and non-enterotoxigenic Staphylococci isolated from goats and cattle. J Chin Soc Anim Sci. 2002, 31: 79-86.
Zhuang LP: Survey of the enterotoxigenic types for staphylococcal isolates obtained from goats, food and clinical samples. Master thesis. 1993, National Chung Hsing University, Department of Food Science and Biotechnology
Vautor E, Abadie G, Guibert JM, Huard C, Pépin M: Genotyping of Staphylococcus aureus isolated from various sites on farms with dairy sheep using pulsed-field gel electrophoresis. Vet Microbiol. 2003, 96: 69-79. 10.1016/S0378-1135(03)00207-4.
Juang ZW: Study of antibiotic resistance and epidemiological typing of Staphylococcus aureus isolated from bovine mastitis in Taiwan. Master Thesis. 2005, National Chung Hsing University, Department of Veterinary Medicine
Zadoks R, van Leeuwen W, Barkema H, Sampimon O, Verbrugh H, Schukken YH, van Belkum A: Application of pulsed-field gel electrophoresis and binary typing as tools in veterinary clinical microbiology and molecular epidemiologic analysis of bovine and human Staphylococcus aureus isolates. J Clin Microbiol. 2000, 38: 1931-1939.
Lange C, Cardoso M, Senczek D, Schwarz S: Molecular subtyping of Staphylococcus aureus isolates from cases of bovine mastitis in Brazil. Vet Microbiol. 1999, 67: 127-141. 10.1016/S0378-1135(99)00031-0.
Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, Hiramatsu K: Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2001, 45: 1323-1336. 10.1128/AAC.45.5.1323-1336.2001.
Davis SL, Rybak MJ, Amjad M, Kaatz GW, McKinnon PS: Characteristics of patients with healthcare-associated infection due to SCCmec type IV methicillin-resistant Staphylococcus aureus. Infect Control Hosp Epidemiol. 2006, 27: 1025-1031. 10.1086/507918.
Cruz FD, Davies J: Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol. 2000, 8: 128-133. 10.1016/S0966-842X(00)01703-0.
Lee JH: Methicillin (Oxacillin)-resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl Environ Microbiol. 2003, 69: 6489-6494. 10.1128/AEM.69.11.6489-6494.2003.
Hsieh JM, Chen RS, Tsai TY, Pan TM, Chou CC: Phylogenetic analysis of livestock oxacillin-resistant Staphylococcus aureus. Vet Microbiol. 2008, 126: 234-242. 10.1016/j.vetmic.2007.07.013.
Chen RS: Analysis of staphlococcal enterotoxins from workers and livestock animals isolates. Master Thesis. 2005, National Taiwan University, Graduate Institute of Veterinary Medicine
Lowy FD: Staphylococcus aureus infections. N Engl J Med. 1998, 339: 520-532. 10.1056/NEJM199808203390806.
Le Loir Y, Baron F, Gautier M: Staphylococcus aureus and food poisoning. Genet Mol Res. 2003, 31: 63-76.
Joo YS, Fox LK, Davis WC, Bohach GA, Park YH: Staphylococcus aureus associated with mammary glands of cows: genotyping to distinguish different strain s among herds. Vet Microbiol. 2001, 80: 131-138. 10.1016/S0378-1135(00)00381-3.
Hsueh PR, Liu YC, Yang D, Yan JJ, Wu TL, Huang WK, Wu JJ, Ko WC, Leu HS, Yu CR, Luh KT: Multicenter surveillance of antimicrobial resistance of major bacterial pathogens in intensive care units in 2000 in Taiwan. Microb Drug Resist. 2001, 7: 373-382. 10.1089/10766290152773383.
Waldner DN: Sample collection and handling. Laboratory handbook on bovine mastiti. Edited by: Hogan JS, Gonzalez RN, Harmon RJ, Nickerson SC, Oliver SP, Pankey JW, Smith KL. 1999, Wisconsin: National Mastitis Council, 1-11.
Davidson TJ, Dohoo IR, Alan WD, Harry H, Kathy C: A cohort study of coagulase negative staphylococcal mastitis in selected dairy herds in Prince Edward Island. Can J Vet Res. 1992, 56: 275-280.
Myllys V, Ridell J, Bjorkroth J, Biese I, Pyoarla S: Persistence in bovine mastitis of Staphylococcus aureus clones as assessed by random amplified polymorphic DNA analysis, biotyping and ribotyping. Vet Microbiol. 1997, 57: 245-251. 10.1016/S0378-1135(97)00137-5.
Clinical and Laboratory Standards Institute: Performance standards for antimicrobial disc susceptibility tests; approved standard-9th ed. Wayne. 2006
Song MD, Wachi M, Doi M, Ishino F, Matsuhashi M: Evolution of an inducible penicillin-target protein in methicillin-resistant Staphylococcus aureus by gene fusion. FEBS Lett. 1987, 221: 167-171. 10.1016/0014-5793(87)80373-3.
Ito T, Katayama Y, Hiramatsu K: Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob Agents Chemother. 1999, 43: 1449-1458.
Betley MJ, Mekalanos JJ: Nucleotide sequence of the type A staphylococcal enterotoxin gene. J Bacteriol. 1988, 170: 34-41.
Jones CL, Khan SA: Nucleotide sequence of the enterotoxin B gene from Staphylococcus aureus. J Bacteriol. 1986, 166: 29-33.
Bohach GA, Schlievert PM: Nucleotide sequence of the staphylococcal enterotoxin C1 gene and relatedness to other pyrogenic toxins. Mol Gen Genet. 1987, 209: 15-20. 10.1007/BF00329830.
Bayles KW, Iandolo JJ: Genetic and molecular analyses of the gene encoding staphylococcal enterotoxin D. J Bacteriol. 1989, 171: 4799-4806.
Couch JL, Soltis MT, Betley MJ: Cloning and nucleotide sequence of the type E staphylococcal enterotoxin gene. J Bacteriol. 1988, 170: 2954-2960.
Munson SH, Tremaine MT, Betley MJ, Welch RA: Identification and characterization of staphylococcal enterotoxin types G and I from Staphylococcus aureus. Infect Immun. 1998, 66: 3337-3348.
Ren K, Bannan JD, Pancholi V, Cheung AL, Robbins JC, Fischetti VA, Zabriskie JB: Characterization and biological properties of a new staphylococcal exotoxin. J Exp Med. 1994, 180: 1675-1683. 10.1084/jem.180.5.1675.
Zhang S, Iandolo JJ, Stewart GC: The enterotoxin D plasmid of Staphylococcus aureus encodes a second enterotoxin determinant (sej). FEMS Microbiol Lett. 1998, 168: 227-233. 10.1111/j.1574-6968.1998.tb13278.x.
Nikolay S, Dmitriy V, Vladimir C, Avraham R: Simultaneous analysis of multiple staphylococcal enterotoxin genes by an oligonucleotide microarray assay. J Clin Microbiol. 2004, 42: 2134-2143. 10.1128/JCM.42.5.2134-2143.2004.
Bannerman TL, Hancock GA, Tenover FC, Miller JM: Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J Clin Microbiol. 1995, 33: 551-555.
Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG: Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000, 38: 1008-1015.
Acknowledgements
The authors would like to thank the grant from the Taiwan Council of Agriculture Executive Yuan of the Republic of China, Taiwan (AS-97-14.2.1-BQ-B1 and AS-98-9.2.4-BQ-B1) and Yunlin and Chiayi Animal Disease Control Center veterinarians assisted with collecting the samples.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
CC instructed the molecular genetics and phylogenetic studies, and edited the manuscript. CYY designed experiments and assisted in data analysis. YHL performed the experiments. YCS coordinated and designed the experiments, and prepared the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Chu, C., Yu, C., Lee, Y. et al. Genetically divergent methicillin-resistant Staphylococcus aureus and sec-dependent mastitis of dairy goats in Taiwan. BMC Vet Res 8, 39 (2012). https://doi.org/10.1186/1746-6148-8-39
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1746-6148-8-39