Abstract
Background
Twenty four non replicate imipenem resistant P. aeruginosa were isolated between January and November 2008, in the kidney transplantation unit of Charles Nicolle Hospital of Tunis (Tunisia). This study was conducted in order to establish epidemiological relationship among them and to identify the enzymatic mechanism involved in imipenem resistance.
Methods
Analysis included antimicrobial susceptibility profile, phenotypic (imipenem-EDTA synergy test) and genotypic detection of metallo-β-lactamase (MBL) (PCR), O-serotyping and pulsed-field gel electrophoresis.
Results
All strains showed a high level of resistance to all antimicrobials tested except to colistin. The presence of MBL showed concordance between phenotypic and genotypic methods. Sixteen isolates were identified as VIM-2 MBL-producers and 13 of them were serotype O4 and belonged to a single pulsotype (A).
Conclusions
This study describes an outbreak of VIM-2-producing P. aeruginosa in a kidney transplantation unit. Clinical spread of blaVIM-2 gene is a matter of great concern for carbapenem resistance in Tunisia.
Similar content being viewed by others
Background
Pseudomonas aeruginosa is a frequent nosocomial pathogen that causes a wide range of opportunistic infections and nosocomial outbreaks [1–3]. Its high intrinsic resistance to antibiotics and ability to develop multidrug resistance pose serious therapeutic problems. Four broad-spectrum β-lactams, such as carbapenems, are potential drugs for the therapy of infections caused by P. aeruginosa[4, 5]. However, the increasing use of these compounds has resulted in the emergence of carbapenem-resistant P. aeruginosa isolates, limiting treatment options [6, 7]. Most carbapenem resistance is due to impermeability, which arises via loss of the OprD (D2) porin, but carbapenem hydrolysing metallo-β-lactamases (MBLs) are increasingly reported [8]. Genes encoding MBLs are located as cassettes in integrons that provide them with the potential for expression and dissemination [9, 10]. To date, nine MBL types, namely, IMP-like[9], VIM-like[9], SPM-1[11], GIM-1[12], SIM-1[13], AIM-1[14], KHM-1[15], NDM-1[16] and DIM-1[17], have been identified in Gram negative bacilli. Worldwide, the IMP and VIM types are the most commonly detected MBLs in P. aeruginosa[9, 10]. VIM-type MBLs are predominant in the Mediterranean region [9, 10].
At Charles Nicolle hospital of Tunisia, since November 2002, VIM-2 producing P. aeruginosa has been isolated, mainly in surgery and intensive care unit [18]. In 2008, an increasing rate of imipenem resistance in P. aeruginosa was observed in the kidney transplantation unit. The aim of the present study was to determine the occurrence of MBL genes among imipenem resistant isolates and to establish epidemiological relationship among them.
Methods
Patients, Bacterial strains, Serotyping and susceptibility testing
Twenty four non duplicate imipenem resistant P. aeruginosa isolates were recovered between January and November 2008 in the kidney transplantation unit of Charles Nicolle hospital of Tunis. They were isolated from urine (n = 20), cutaneous pus (n = 3) and blood (n = 1).
All samples were taken for microbial diagnosis from 24 different patients [sex ratio 3.8]. The median age of the patients was 51 years [range: 34 to 80 years]. They were admitted in the urology unit (n = 10) and nephrology unit (n = 14).
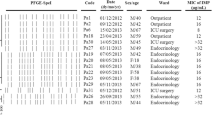
A retrospective review was conducted for only 15 patients (6 from urology unit, 9 from the nephrology unit). Date of transplantation was mentioned in Table 1. However, no data concerning kind of transplantation and live donator was collected. All patients presented chronic renal failure associated in 13 cases to at least one underlying co-morbidity [High blood pressure (n = 13), systemic lupus erythematosis (n = 1), diabetes (n = 1), chronic glomerulonephritis (n = 5), nephritic syndrome (n = 1) and renal tuberculosis (n = 1)]. In all patients fever was the major symptom. Ceftazidime combination therapy with colistin accounted for all patients. Attributable mortality was 5% of cases.
Bacterial identification was performed using ApiNE (bio-Mérieux, Marcy-l'Etoile, France). O-serotyping was determined by slide agglutination test using polyvalent antisera and 16 monovalent antisera numbered O1-O16 according to the manufacturer's instructions (Bio-Rad, Marnes-La-Coquette, France).
Antimicrobial susceptibility was tested with the agar disk diffusion method according to the CLSI guidelines [19]. The MICs of ticarcillin, ticarcillin + clavulanic acid, ceftazidime, aztreonam, cefepime, imipenem and meropenem were determined using the dilution method in Mueller Hinton agar according to the CLSI guidelines [19]. E. coli ATCC 25922 and P. aeruginosa ATCC 27853 were used as control strains.
To detect MBL production, the imipenem-EDTA synergy test was used [20]. An enlargement of the inhibition zone of imipenem facing the disc of EDTA was considered as a positive test.
PCR amplification
For the detection of blaVIM and blaIMP genes PCR experiments were performed using consensus primers as described previously [21]. Primers specific for blaVIM-2 were also used [22].
Clonal relationship by Pulsed-Field Gel Electrophoresis (PFGE)
Molecular typing of MBL producing isolates was carried out, as described previously by Pulsed-Field Gel Electrophoresis (PFGE) using Spe I restriction endonuclease [23]. Clonal relationships based on PFGE patterns were interpreted according to the criteria proposed by Tenover et al [24].
Results and discussion
Carbapenems are potent agents against multiresistant Gram negative bacilli, including P. aeruginosa, but their efficacy is increasingly compromised by the emergence and the worldwide dissemination of carbapenem resistance strains, which are implicated in large outbreaks as described in many countries [1, 25]. In Tunisia, frequencies of imipenem resistant P. aeruginosa varies between 16% and 37.6% [26–28]. At Charles Nicolle Hospital of Tunis, their frequency was stable until 2004 (1%), but increased dramatically from 2005 (25%). They were mainly isolated in surgery and intensive care unit [18], but in 2008, an increasing rate of multidrug-resistant P. aeruginosa was observed in the kidney transplantation unit. All strains exhibited a multidrug-resistant phenotype; they were resistant to antipseudomonal β-lactams (including aztreonam), aminoglycosides and fluoroquinolones. They remained susceptible only to colistin which is used for the treatment of our kidney transplanted patients despite its renal toxicity [29]. MICs results are shown in table 1. All strains showed a high level of resistance to all β-lactams, particularly to carbapenems (> 512 μg/ml). Only 16 strains (67%) were positive according to the imipenem-EDTA synergy test, suggesting the presence of MBLs. The acquisition of a MBL gene alone does not necessarily confer elevated level of resistance to carbapenems. Indeed, secondary changes in regulatory system of MBL gene expression, outer membrane permeability, active efflux systems in bacterial membrane, and/or multiplication of structure gene might well be implicated in acquisition of high-level carbapenem resistance [30, 31]. Aztreonam is the only β-lactam that may remain fully active against MBL producers [8], however all our strains were resistant to this β-lactam, suggesting the occurrence of other mechanisms of β-lactam resistance [32]. The eight MBL negative strains were also resistant to all antibiotic tested (Table 1), but the mechanism involved in the resistance has not been further examined.
The 16 MBLs-producer strains were positive for bla VIM-2 gene and none strain harboured the blaIMP gene (Table 1). In Tunisia, the most common MBL identified VIM-2 in accordance with the actual situation worldwide [9, 33, 34]. Historically, the first reports of MBLs genes were VIM-2 and VIM-4 types in P. aeruginosa[18, 35] and K. pneumoniae[36] respectively.
Serotyping identified 3 serotypes: O4 (n = 16), O11 (n = 4) and O12 (n = 1) (table 1). Only 2 strains were nontypeable with monovalent antisera. In Tunisia [37] as well as in many European countries [38], it has been repeatedly demonstrated over the past 20 years that serotypes O11 and O12 dominate among multiresistant P. aeruginosa isolates. The 16 MBL-positive strains were divided into 3 pulsotypes designed A (n = 13), B (n = 2) and C (n = 1) (Table 1). The 13 strains of pulsotype A were of serotype O4. These results imply that the dissemination of VIM-2 in our kidney transplantation unit was mainly due to the of spread clonal strains, however, unrelated VIM-2-harboring strains occurred. Outbreaks of VIM β-lactamase-producing P. aeruginosa have been also reported in Greece [3], Italy [2] and Kenya [39], but there is still very limited knowledge on the epidemiology of MBLs in Africa.
The emergence of acquired MBLs among P. aeruginosa represents an epidemiological risk for at least two reasons: firstly, MBLs confer resistance not only to carbapenems but to virtually all β-lactams and are frequently associated with resistance to aminoglycosides; and secondly, genes encoding for MBL enzymes are most commonly carried on mobile genetic elements (integrons, plasmids, transposons) that can spread horizontally among unrelated strains [9, 18].
Indeed, the blaVIM-2 gene was found in strains of different genotypes (A, B and C), reflecting its ability to transfer from one bacterium to another.
Conclusion
In conclusion, we found an outbreak of VIM-2-producing P. aeruginosa in a kidney transplantation unit. Thus, blaVIM-2 gene may have been spreading in Gram negative rod in Tunisia. This emphasizes the necessity of early recognition of MBL producing isolates, rigorous infection control, and restricted clinical use of broad-spectrum β-lactams including carbapenems.
Abbreviations
- MBL:
-
metallo-beta-lactamase
- PFGE:
-
Pulsed-Field Gel Electrophoresis.
References
Kohlenberg A, Weitzel-Kage D, van der Linden P, Sohr D, Vogeler S, Kola A, Halle E, Ruden H, Weist K: Outbreak of carbapenem-resistant Pseudomonas aeruginosa infection in a surgical intensive care unit. J Hosp Infect. 2010, 74: 350-357. 10.1016/j.jhin.2009.10.024.
Cornaglia G, Mazzariol A, Lauretti L, Rossolini GM, Fontana R: Hospital outbreak of carbapenem-resistant Pseudomonas aeruginosa producing VIM-1, a novel transferable metallo-β-lactamase. Clin Infect Dis. 2000, 31: 1119-1125. 10.1086/317448.
Tsakris A, Pournaras S, Woodford N, Palepou MF, Babini GS, Douboyas J, Livermore DM: Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J Clin Microbiol. 2000, 38: 1290-1292.
Aloush V, Navon-Venezia S, Seigman-Igra Y, Cabili S, Carmeli Y: Multidrug-resistant Pseudomonas aeruginosa: risk factors and clinical impact. Antimicrob Agents Chemother. 2006, 50: 43-48. 10.1128/AAC.50.1.43-48.2006.
Rossolini GM, Mantengoli E: Treatment and control of severe infections caused by multiresistant Pseudomonas aeruginosa. Clin Microbiol Infect. 2005, 11 (Suppl 4): 17-32.
Falagas ME, Kopterides P: Risk factors for the isolation of multi-drug-resistant Acinetobacter baumannii and Pseudomonas aeruginosa: a systematic review of the literature. J Hosp Infect. 2006, 64: 7-15. 10.1016/j.jhin.2006.04.015.
Zavascki AP, Cruz RP, Goldani LZ: Risk factors for imipenem-resistant Pseudomonas aeruginosa: a comparative analysis of two case-control studies in hospitalized patients. J Hosp Infect. 2005, 59: 96-101. 10.1016/j.jhin.2004.09.007.
Livermore DM, Woodford N: Carbapenemases: a problem in waiting?. Curr Opin Microbiol. 2000, 3: 489-495. 10.1016/S1369-5274(00)00128-4.
Walsh TR, Toleman MA, Poirel L, Nordmann P: Metallo-β-lactamases: the quiet before the storm?. Clin Microbiol Rev. 2005, 18: 306-325. 10.1128/CMR.18.2.306-325.2005.
Queenan AM, Bush K: Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev. 2007, 20: 440-458. 10.1128/CMR.00001-07.
Murphy TA, Simm AM, Toleman MA, Jones RN, Walsh TR: Biochemical characterization of the acquired metallo-β-lactamase SPM-1 from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2003, 47: 582-587. 10.1128/AAC.47.2.582-587.2003.
Castanheira M, Toleman MA, Jones RN, Schmidt FJ, Walsh TR: Molecular characterization of a β-lactamase gene, bla GIM-1 , encoding a new subclass of metallo-β-lactamase. Antimicrob Agents Chemother. 2004, 48: 4654-4661. 10.1128/AAC.48.12.4654-4661.2004.
Lee K, Yum JH, Yong D, Lee HM, Kim HD, Docquier JD, Rossolini GM, Chong Y: Novel acquired metallo-beta-lactamase gene, bla(SIM-1), in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob Agents Chemother. 2005, 49: 4485-4491. 10.1128/AAC.49.11.4485-4491.2005.
Gupta V: Metallo beta lactamases in Pseudomonas aeruginosa and Acinetobacter species. Expert Opin Investig Drugs. 2008, 17: 131-143. 10.1517/13543784.17.2.131.
Sekiguchi J, Morita K, Kitao T, Watanabe N, Okazaki M, Miyoshi-Akiyama T, Kanamori M, Kirikae T: KHM-1, a novel plasmid-mediated metallo-beta-lactamase from a Citrobacter freundii clinical isolate. Antimicrob Agents Chemother. 2008, 52: 4194-4197. 10.1128/AAC.01337-07.
Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR: Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009, 53: 5046-5054. 10.1128/AAC.00774-09.
Poirel L, Rodriguez-Martinez JM, Al Naiemi N, Debets-Ossenkopp YJ, Nordmann P: Characterization of DIM-1, an integron-encoded metallo-beta-lactamase from a Pseudomonas stutzeri clinical isolate in the Netherlands. Antimicrob Agents Chemother. 2010, 54: 2420-2424. 10.1128/AAC.01456-09.
Hammami S, Gautier V, Ghozzi R, Da Costa A, Ben-Redjeb S, Arlet G: Diversity in VIM-2-encoding class 1 integrons and occasional blaSHV2a carriage in isolates of a persistent, multidrug-resistant Pseudomonas aeruginosa clone from Tunis. Clin Microbiol Infect. 2010, 16: 189-193. 10.1111/j.1469-0691.2009.03023.x.
Clinical and Laboratory Standards Institute: Performance Standards for Antimicrobial Susceptibility Testing. Sixteenth information supplement. M100-S16. 2006, Wayne, PA: CLSI
Pitout JD, Chow BL, Gregson DB, Laupland KB, Elsayed S, Church DL: Molecular epidemiology of metallo-β-lactamase-producing Pseudomonas aeruginosa in the Calgary Health Region: emergence of VIM-2-producing isolates. J Clin Microbiol. 2007, 45: 294-298. 10.1128/JCM.01694-06.
Yatsuyanagi J, Saito S, Ito Y, Ohta K, Kato J, Harata S, Suzuki N, Amano K: Identification of Pseudomonas aeruginosa clinical strains harboring the bla VIM-2 metallo-β-lactamase gene in Akita Prefecture, Japan. Jpn J Infect Dis. 2004, 57: 130-132.
Yan JJ, Hsueh PR, Ko WC, Luh KT, Tsai SH, Wu HM, Wu JJ: Metallo-β-lactamases in clinical Pseudomonas isolates in Taiwan and identification of VIM-3, a novel variant of the VIM-2 enzyme. Antimicrob Agents Chemother. 2001, 45: 2224-2228. 10.1128/AAC.45.8.2224-2228.2001.
Patzer JA, Dzierzanowska D: Increase of imipenem resistance among Pseudomonas aeruginosa isolates from a Polish paediatric hospital (1993-2002). Int J Antimicrob Agents. 2007, 29: 153-158. 10.1016/j.ijantimicag.2006.08.044.
Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B: Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995, 33: 2233-2239.
Cezario RC, Duarte De Morais L, Ferreira JC, Costa-Pinto RM, da Costa Darini AL, Gontijo-Filho PP: Nosocomial outbreak by imipenem-resistant metallo-beta-lactamase-producing Pseudomonas aeruginosa in an adult intensive care unit in a Brazilian teaching hospital. Enferm Infecc Microbiol Clin. 2009, 27: 269-274. 10.1016/j.eimc.2008.09.009.
Kalai S, Achour W, Abdeladhim A, Bejaoui M, Ben Hassen A: Pseudomonas aeruginosa isolés de patients immunodéprimés: résistance aux antibiotiques, sérotypage et typage moléculaire. Med Mal Infect. 2005, 35: 530-535. 10.1016/j.medmal.2005.08.006.
Ben Abdallah H, Noomen S, Khelifa AB, Sahnoun O, Elargoubi A, Mastouri M: Susceptibility patterns of Pseudomonas aeruginosa strains isolated in the Monastir region, Tunisia. Med Mal Infect. 2008, 38: 554-556. 10.1016/j.medmal.2008.05.002.
Lamia T, Bousselmi K, Saida BR, Allah MA: Epidemiological profile and antibiotic susceptibility of Pseudomonas aeruginosa isolates within the burned patient hospitalized in the intensive care burn unit. Tunis Med. 2007, 85: 124-127.
Evans ME, Feola DJ, Rapp RP: Polymyxin B sulfate and colistin: old antibiotics for emerging multiresistant gram-negative bacteria. Ann Pharmacother. 1999, 33: 960-967. 10.1345/aph.18426.
Livermore DM: Of Pseudomonas, porins, pumps and carbapenems. J Antimicrob Chemother. 2001, 47: 247-250. 10.1093/jac/47.3.247.
Rodriguez-Martinez JM, Poirel L, Nordmann P: Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009, 53: 4783-4788. 10.1128/AAC.00574-09.
Lolans K, Queenan AM, Bush K, Sahud A, Quinn JP: First nosocomial outbreak of Pseudomonas aeruginosa producing an integron-borne metallo-β-lactamase (VIM-2) in the United States. Antimicrob Agents Chemother. 2005, 49: 3538-3540. 10.1128/AAC.49.8.3538-3540.2005.
Toleman MA, Biedenbach D, Bennett DM, Jones RN, Walsh TR: Italian metallo-β-lactamases: a national problem? Report from the SENTRY Antimicrobial Surveillance Programme. J Antimicrob Chemother. 2005, 55: 61-70.
Poirel L, Lambert T, Turkoglu S, Ronco E, Gaillard J, Nordmann P: Characterization of Class 1 integrons from Pseudomonas aeruginosa that contain the bla VIM-2 carbapenem-hydrolyzing β-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob Agents Chemother. 2001, 45: 546-552. 10.1128/AAC.45.2.546-552.2001.
Mansour W, Poirel L, Bettaieb D, Bouallegue O, Boujaafar N, Nordmann P: Metallo-beta-lactamase-producing Pseudomonas aeruginosa isolates in Tunisia. Diagn Microbiol Infect Dis. 2009, 64: 458-461. 10.1016/j.diagmicrobio.2009.04.003.
Ktari S, Arlet G, Mnif B, Gautier V, Mahjoubi F, Ben Jmeaa M, Bouaziz M, Hammami A: Emergence of multidrug-resistant Klebsiella pneumoniae isolates producing VIM-4 metallo-β-lactamase, CTX-M-15 extended-spectrum β-lactamase, and CMY-4 AmpC β-lactamase in a Tunisian university hospital. Antimicrob Agents Chemother. 2006, 50: 4198-4201. 10.1128/AAC.00663-06.
Hammami S, Ghozzi R, Ben Ayed S, Ben Hassen A, Ben Rejeb S: Clonal spread of carbapenem resistant Pseudomonas aeuriginosa in an university hospital. Tunis Med. 2008, 86: 653-656.
Libisch B, Watine J, Balogh B, Gacs M, Muzslay M, Szabo G, Fuzi M: Molecular typing indicates an important role for two international clonal complexes in dissemination of VIM-producing Pseudomonas aeruginosa clinical isolates in Hungary. Res Microbiol. 2008, 159: 162-168. 10.1016/j.resmic.2007.12.008.
Pitout JD, Revathi G, Chow BL, Kabera B, Kariuki S, Nordmann P, Poirel L: Metallo-β-lactamase-producing Pseudomonas aeruginosa isolated from a large tertiary centre in Kenya. Clin Microbiol Infect. 2008, 14: 755-759. 10.1111/j.1469-0691.2008.02030.x.
Acknowledgements
This work was financed by grants from the Ministry of Scientific Research, Technology and Competence Development of Tunisia.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SH designed the study and wrote the manuscript. IBBB, RG, MS, SA and SBR performed critical reading of manuscript and supervision. All authors have read and approved the final manuscript.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Hammami, S., Boutiba-Ben Boubaker, I., Ghozzi, R. et al. Nosocomial outbreak of imipenem-resistant Pseudomonas aeruginosa producing VIM-2 metallo-β-lactamase in a kidney transplantation unit. Diagn Pathol 6, 106 (2011). https://doi.org/10.1186/1746-1596-6-106
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1746-1596-6-106