Abstract
Upon cell infection by a retrovirus, the viral DNA polymerase, called reverse transcriptase (RT), copies the genomic RNA to generate the proviral DNA flanked by two long terminal repeats (LTR). A discovery twenty years ago demonstrated that the structural viral nucleocapsid protein (NC) encoded by Gag is an essential cofactor of reverse transcription, chaperoning RT during viral DNA synthesis. However, it is only recently that NC was found to exert a control on the timing of reverse transcription, in a spatio-temporal manner. This brief review summarizes findings on the timing of reverse transcription in wild type HIV-1 and in nucleopcapsid (NC) mutants where virions contain a large amount of newly made viral DNA. This brief review also proposes some explanations of how NC may control late reverse transcription during Gag assembly in virus producer cells.
Similar content being viewed by others
Review
Overview on the process of viral DNA synthesis by RT
Retroviruses differ from other positive-strand RNA viruses in the sense that their genomic RNA is reverse transcribed to generate a double stranded DNA flanked by two long terminal repeats (LTR). This essential multistep process is performed by the retroviral RNA/DNA dependent DNA polymerase named reverse transcriptase (RT) discovered in 1970 [1–3]. Originally, the RT activity was found in purified avian and murine virions upon treatment with a low concentration of a non-ionic detergent [1–6]. At almost the same time, RT activity was also found in virus-like particles purified from human fluids and cells [7, 8]. Later, an RT-associated RNaseH activity was characterized and found to be relevant to the process of viral DNA synthesis [9, 10]. The series of reactions carried out by RT to copy the retroviral genome in order to generate the double stranded viral DNA which is then integrated into the host-cell genome has been known for almost 25 years. Reverse transcription first requires a specific cellular tRNA annealed to the primer binding site (PBS) for the initiation of cDNA synthesis (Figure 1) and two obligatory DNA strand transfers to carry out the synthesis of the complete, LTR flanked, proviral DNA [11–13]. The dimeric nature of the retroviral RNA genome is largely responsible for the high genetic variability of highly replicating viruses such as Rous sarcoma virus (RSV) and HIV-1 by means of forced and non-forced copy-choice recombinations during reverse transcription [12, 14, 15].
A scheme of the HIV-1 replication complex. The genomic RNA in a dimeric form is coated by about 1500 nucleocapsid (NC) protein molecules (in red) in the viral core particle. Molecular interactions have been characterized, at least in part, namely (i) the two viral RNAs via DIS, DLS, 5'-3' and other interactions to form the dimeric 60S complex (black lines), (ii) the primer tRNALys3 (thin black line) annealed to the viral PBS RNA, (iii) NC molecules coating the viral RNA and tRNA (NC basic residues and zinc fingers), (iv) the primer tRNALys3 bound to RTp66-p51(in blue), (v) the viral RNA and RT, (vi) NC and RT and (vii) NC and NC molecules. Viral proteins such as IN and Vif that may play a role in viral DNA synthesis are not represented in this scheme. For references see text.
The discovery of HIV-1 and the AIDS epidemic fueled unprecedent interest in and support for basic research on retroviruses as well as extensive efforts aimed at combating the AIDS virus. As a result, the structure-function relationships of HIV-1 RT have been and continue to be intensively studied using a multidisciplinary approach. The 3D structure of RT in its p66-p51 free form was established [15–17], and more recently the specific orientation of the RT polymerase and RNaseH active sites was characterized using single molecule assays in vitro [18, 19]. Later, it was discovered that the major virion protein of the inner core of alpha and gamma-retroviruses and lentiviruses, the nucleocapsid protein (NC) encoded by Gag was a key cofactor of the RT enzyme, chaperoning obligatory steps in viral DNA synthesis [20–28]. At the same time, the NC domain of the Gag structural polyprotein was found to direct genomic RNA selection, packaging and dimerization during virion assembly [29–34]. Thus NC is a multifunctional virus structural protein necessary for the completion of the early and late phases of retrovirus replication (reviewed in [28, 35–38]).
How then can we explain the multiple roles of NC? NC is a potent nucleic acid chaperone, which tightly binds nucleic acids and facilitates the annealing of complementary sequences as well as strand transfer and exchange reactions in physiological conditions (reviewed in [35–38]). NC is encoded by Gag in most, if not all, retroviruses and retrotransposons [36] where its unique chaperoning activity ensures primer tRNA annealing to the genomic PBS and the obligatory minus and plus DNA strand transfers that are required for the synthesis of the complete, LTR flanked, viral DNA [28, 37, 38].
In the inner core of the HIV-1 viral particle, about 1500 NC molecules [39] coat the genomic RNA in the form of NC oligomers [28, 40]. In addition, tight interactions were found to take place between NC molecules, the cellular tRNA primer and the RT enzyme in reconstituted HIV-1 replicative complexes (Figure 1). These multiple RT-NC-RNA interactions contribute to the fidelity of the reverse transcripton reaction by inhibiting self-initiation of cDNA synthesis and providing excision-repair activities to the RT enzyme in vitro [24, 39, 41–46].
After cell infection, the virion core is released into the cytoplasm where it is believed to undergo structural alterations giving rise to a large ribonucleoprotein structure called the reverse transcription complex (RTC), the site of extensive viral DNA synthesis. The RTC is thought to predominantly consist of the genomic RNA coated with NC protein molecules and other components such as the matrix (MA), capsid (CA) and viral protein R (Vpr) molecules together with the viral enzymes RT and integrase (IN) [47–49]. A different view was recently provided by biochemical and electron microscopy studies showing that HIV-1 cores remained in the cytoplasm of newly infected cells up to the nuclear pore [50–54]. These results strongly suggest that completion of proviral DNA synthesis most probably relies on the proper structure and the stability of the viral cores.
Newly made viral DNA in retroviral particles
The canonical view of retrovirus formation, notably that of HIV-1, states that the overall process takes place at the plasma membrane where Gag molecules assemble via interactions between MA and the phospholipids on the one hand, and between NC and the genomic RNA on the other (reviewed in [55]). Upon completion, immature particles are produced by budding during which Gag and Pol processing by the viral protease (PR) occurs, ultimately leading to the condensation of the inner core [32, 56]. However, a series of results indicate that assembly can also take place on intracellular membranes such as endosomes and multivesicular bodies [57–60]. The PR enzyme may therefore already be active at the onset of assembly directing the cleavage of the Gag and Gag-Pol polyproteins, as evidenced by the presence of mature CA, MA and NC proteins in cytoplasmic extracts of infected cells. In both cases, the newly made viral particles are thought to contain the full length viral RNA in a dimeric form as the genetic material along with minor quantities of spliced RNAs [61].
However, small amounts of viral cDNA were found in newly made viral particles of RSV, Moloney murine leukemia virus (MoMLV), and HIV-1 indicating that an active RT enzyme can function during virus assembly [10, 62–64]. This notion of premature reverse transcription has been confirmed by Zhang et al. [64] who showed that AZT treatment of HIV-1 infected T cells resulted in a 10–100 fold decrease of the intravirion cDNA level. In addition, the physiological microenvironment, for example the seminal fluid, was found to enhance the accumulation of intravirion viral DNA by a process called natural endogenous reverse transcription (NERT) [64]. Interestingly, synthesis of a full length infectious viral DNA can be achieved in virions of MLV and equine infectious anemia virus (EIAV) under well defined in vitro conditions [6, 65], that probably reconstitute the microenvironment promoting extensive NERT, especially components present in the seminal fluid [66].
The newly synthesized viral DNA present in infecting virions was shown to play a key role in vivo because it augments virus infection of non-activated human primary target cells by nearly one hundred fold while it has no effect on activated T cells [66]. The role of the physiological microenvironment is not limited to viral DNA synthesis since a recently identified aggregating prostatic acidic phosphatase (PAP)-derived peptide that is abundant in the seminal fluid was shown to augment virus to cell attachment and entry, thus facilitating the very early event of HIV-1 infection during a sexual intercourse [67, 68].
HIV-1 NC and the timing of reverse transcription
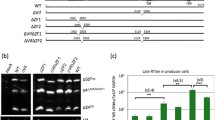
It has long been shown that mutating the highly conserved CCHC residues of the NC zinc fingers impairs genomic RNA packaging and results in the production of replication defective viral particles (reviewed in [28, 37, 38]). Moreover, mutations affecting the 3D structure of the zinc fingers or their respective orientation cause a decrease in the genomic RNA content of HIV-1 viral particles and result in the production of defective particles, although significant amounts of viral DNA can be synthesized in infected cells ([26, 27, 45, 69]; reviewed in [37, 38, 70]). However, this has been recently disputed. In fact, the influence of NC mutations on unsuspected aspects of HIV-1 virion formation has just been discovered, in particular the influence on packaging of multispliced 1.8 kb RNA (MS RNA) and NERT [71, 72]. It was found that deleting or mutating the NC zinc fingers or the N-terminal basic residues caused a 10–30 fold reduction of genomic RNA in newly made virions (Fig. 2A), while this had no or a slightly positive influence on virion incorporation of MS RNA that contain part of the packaging Psi element recognized by NC (Fig. 2A). At the same time there was a 10–100 fold enhancement of newly synthesized viral DNA in these NC mutant HIV-1 virions as compared with wild type virions (Fig. 2B; [72, 73]). This unexpected accumulation of viral DNA in NC mutant virions was independently reported by R. Gorelick and colleagues [74].
Nucleic acids content of wild-type and NC mutant HIV-1 virions. WT, ΔZF1, ΔZF2 and ΔZF1ZF2 represent wild type, and deletion of the first, the second and of both CCHC zinc fingers, respectively. All values were determined by quantitative RT-PCR (A) and PCR (B) (n = 3 ± SD). A- Viral RNAs corresponding to the full length (FL) and the multispliced (MS) forms. B- Viral DNAs corresponding to the strong stop (ss), Gag, full length (FL) and mutispliced (MS) forms.
This reverse transcription takes place in virus producer cells since the addition of the RT inhibitor AZT prevented accumulation of viral DNA in virions, in agreement with the earlier findings of Pomerantz et al. [66] on wild type HIV-1 virions. A closer examination of the viral DNA made in these NC mutant virions reveals that large viral DNA fragments accumulated (Fig. 2B) together with cDNA generated by reverse transcription of the incorporated multispliced viral RNAs. A fraction of this viral DNA was found to be functional since it directed Tat synthesis and LTR activation in human cells [72].
These findings not only confirm the key role of NC in RT-directed viral DNA synthesis and probably its maintenance ([43, 45]; reviewed in [70]) but also indicate that NC exerts a control on the timing of reverse transcription. How then can we explain this extensive reverse transcription already in cells producing the HIV-1 NC mutant virions? Within HIV-1 virions with mutations in or deletion of the NC CCHC zinc fingers, the core is formed of mature Gag proteins but it is mostly globular and does not adopt a condensed cone-shaped structure as seen by electron microscopy [75]. These results favor the notion that these NC mutations cause a defect in the late step of Gag assembly. The fact that NC ZF mutants have lost, at least in part, their high binding affinity for the viral RNA [76] and also for the LTR DNA ends [77] could explain the condensation defect of the inner core structure and degradation of the LTR DNA ends [45]. Such ZF mutations also have a negative impact on NC-RT interactions in vitro [42, 77] while the chaperoning activities of the NC mutant proteins either are not or are slightly affected ([78]; reviewed in [37]). The study of Gorelick et al. [74] also showed that mutating the PTAP motif in the Gag C-terminal p6 domain caused a budding delay probably due to a loosening of the interactions between Gag and the cellular transporter protein TSG101 and at the same time led to a strong activation of premature reverse transcription.
Taken together, these findings support the notion that the highly conserved CCHC zinc fingers of NC control formation of a dense core structure where reverse transcription is prevented, at least, partially [72–74]. Thus, the viral NC protein would exert a control on the timing of viral DNA synthesis by the active RT enzyme, delaying the start phase in virus producer cells and chaperoning the entire process until completion in newly infected cells (Figure 3). There are also indications that the conserved CCHC zinc fingers of NC contribute to the maintenance of the complete viral DNA and its IN-mediated integration into the host genome during the course of cell infection [45, 79, 80].
A proposed mechanism for late reverse transcription in cells producing HIV-1 NC mutant particles. A- Newly made Gag and GagPol molecules (1) assemble using the genomic RNA and cellular membrane as platforms, (2) then wild type HIV-1 virions are produced by budding. These processes are facilitated by interactions between NC and cellular proteins (large black arrows). The core containing the genomic RNA is condensed with a cone-shaped structure. A limited level of viral DNA synthesis in producer cells and also anatural endogenous reverse transcription (NERT) can occur (see text). B- In cells producing HIV-1 Gag and GagPol with mutation in or deletion of the nucleocapsid CCHC zinc finger (1), assembly (2) and budding (grey arrow) are probably slowed down due to impaired interactions between NC and the viral RNA. They result in a partial delocalization of Gag in producer cells and a reduced level of newly made viral particles (grey arrow) (see text for references). The resulting virion core is formed of mature Gag proteins, but it is poorly condensed as seen by electron microscopy (see text for references). Taken together, these observations favor the notion that the NC CCHC mutations modify the kinetics of viral assembly which prevent core condensation and could explain, at least in part, why late-premature reverse transcription can readily take place in producer cells.
Future prospects
One outstanding avenue of research is to better understand, at the molecular level, the multiple interactions between NC, RT, and the viral nucleic acids that ensure fidelity and completion of viral DNA synthesis (reviewed in [70]). It is noteworthy that these molecular interactions (Fig. 1) are probably involved in recombinations, by forced and unforced DNA strand transfers via the NC-chaperoning activity, that fuel the genetic diversity of the progeny virus [12, 14]. The zinc fingers and the RT palm domains are probably required for NC-RT recognition [42–45, 81]; yet, the exact interacting domains remain to be determined in vitro and in the viral context.
The physiological microenvironment appears to greatly facilitate HIV-1 NERT. In fact, seminal plasma was found to activate NERT, virus-cell attachment, and entry [64, 66–68]. The presence of molecules such as dNTP, polyamines and a prostatic acidic phosphatase-derived peptide called SEVI in seminal fluid would be responsible, at least in part, for the strong enhancement of HIV-1 infectivity on non-activated primary target cells (op.cit). Yet, the molecular and biochemical mechanisms of such an enhancement in virus infectivity remain to be determined.
A further important avenue of research involves the characterization of compounds aimed at inhibiting HIV-1 RT-NC interactions, and thus the chaperoning [82] of reverse transcription in virus producer cells and in the reverse transcription complex (RTC) in newly infected cells [83]. This work is presently ongoing.
Loosening the interactions between NC protein molecules, the genomic RNA and the RT enzyme by mutating the NC zinc fingers activates viral DNA synthesis in HIV-1 producer cells and results in the production of DNA-containing particles [72–74], similar in nature to foamy viruses (HFV, SFV) where viral DNA synthesis occurs in virus producer cells [84–86]. This finding raises several questions on the nature of functional RT-NC-RNA interactions in most, if not all, retroviruses, notably alpha- and gammaretroviruses such as ASLV and MLV's that contain a mature NC protein. For Foamy viruses, the situation appears much different since RT would have to interact with Gag since it is not itself processed as it is in NC-containing retroviruses. Moreover, no NC-like chaperoning activity has yet been characterized. The structure of HFV/SFV Gag resembles that of Gag of the fruit fly retrovirus Gypsy where the C-terminal domain has NC-like RNA chaperoning activities, but lacks a zinc finger [87]. Thus it would be interesting to examine the putative interaction, if any, between the C-terminal NC-like domain of Gypsy and Foamy virus Gag with the homologous RT enzyme.
Endogenous retroviruses and retrotransposons form a large fraction (42%) of the mammalian genome, but only a very small fraction (<1%) of these endogenous retroviral sequences is expressed and capable of producing viral particles that can be infectious [88, 89]. Nonetheless, the continuous expression of these retroviral sequences in their natural form [90, 91] or as reconstructed, highly active recombinants [92, 93] could be detrimental to the host, possibly causing a variety of diseases [94–97]. Thus endogenous retroviruses such as Gypsy, MLV and HERV which can undergo retrotransposition at a low frequency and behave similarly to exogenous retroviruses would represent interesting model systems to study the spatio-temporal control of viral DNA synthesis by cellular factors [98–100].
References
Temin HM, Mizutani S: RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970, 226 (5252): 1211-3.
Baltimore D: RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970, 226 (5252): 1209-11.
Mizutani S, Boettiger D, Temin HM: A DNA-depenent DNA polymerase and a DNA endonuclease in virions of Rous sarcoma virus. Nature. 1970, 228 (5270): 424-7.
Taylor JM, Faras AJ, Varmus HE, Levinson WE, Bishop JM: Ribonucleic acid directed deoxyribonucleic acid synthesis by the purified deoxyribonucleic acid polymerase of Rous sarcoma virus. Characterization of the enzymatic product. Biochemistry. 1972, 11 (12): 2343-51.
Faras AJ, Taylor JM, McDonnell JP, Levinson WE, Bishop JM: Purification and characterization of the deoxyribonucleic acid polymerase associated with Rous sarcoma virus. Biochemistry. 1972, 11 (12): 2334-42.
Rothenberg E, Smotkin D, Baltimore D, Weinberg RA: In vitro synthesis of infectious DNA of murine leukaemia virus. Nature. 1977, 269 (5624): 122-6.
Schlom J, Spiegelman S, Moore D: RNA-dependent DNA polymerase activity in virus-like particles isolated from human milk. Nature. 1971, 231 (5298): 97-100.
Schlom J, Spiegelman S: Simultaneous detection of reverse transcriptase and high molecular weight RNA unique to oncogenic RNA viruses. Science. 1971, 174 (11): 840-3.
Mölling K, Bolognesi DP, Bauer H, Büsen W, Plassmann HW, Hausen P: Association of viral reverse transcriptase with an enzyme degrading the RNA moiety of RNA-DNA hybrids. Nat New Biol. 1971, 234 (51): 240-3.
Darlix JL, Bromley PA, Spahr PF: New procedure for the direct analysis of in vitro reverse transcription of Rous sarcoma virus RNA. J Virol. 1977, 22 (1): 118-29.
Gilboa E, Mitra SW, Goff S, Baltimore D: A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979, 18 (1): 93-100.
Coffin JM: Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J Gen Virol. 1979, 42 (1): 1-26.
Goff S, Traktman P, Baltimore D: Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J Virol. 1981, 38 (1): 239-48.
Temin HM: Sex and recombination in retroviruses. Trends Genet. 1991, 7 (3): 71-4.
Jacobo-Molina A, Arnold E: HIV reverse transcriptase structure-function relationships. Biochemistry. 1991, 30 (26): 6351-6.
Rodgers DW, Gamblin SJ, Harris BA, Ray S, Culp JS, Hellmig B, Woolf DJ, Debouck C, Harrison SC: The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1995, 92 (4): 1222-6.
Ding J, Das K, Hsiou Y, Sarafianos SG, Clark AD, Jacobo-Molina A, Tantillo C, Hughes SH, Arnold E: Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 A resolution. J Mol Biol. 1998, 284 (4): 1095-111.
Liu S, Abbondanzieri EA, Rausch JW, Le Grice SF, Zhuang X: Slide into action: dynamic shuttling of HIV reverse transcriptase on nucleic acid substrates. Science. 2008, 322 (5904): 1092-7.
Abbondanzieri EA, Bokinsky G, Rausch JW, Zhang JX, Le Grice SF, Zhuang X: Dynamic binding orientations direct activity of HIV reverse transcriptase. Nature. 2008, 453 (7192): 184-9.
Prats AC, Sarih L, Gabus C, Litvak S, Keith G, Darlix JL: Small finger protein of avian and murine retroviruses has nucleic acid annealing activity and positions the replication primer tRNA onto genomic RNA. EMBO J. 1988, 7 (6): 1777-83.
Barat C, Lullien V, Schatz O, Keith G, Nugeyre MT, Grüninger-Leitch F, Barré-Sinoussi F, LeGrice SF, Darlix JL: HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. EMBO J. 1989, 8 (11): 3279-85.
Allain B, Lapadat-Tapolsky M, Berlioz C, Darlix J-LL: Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 1994, 13 (4): 973-81.
Tsuchihashi Z, Brown PO: DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1994, 68 (9): 5863-70.
Guo J, Henderson LE, Bess J, Kane B, Levin JG: Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J Virol. 1997, 71 (7): 5178-88.
Guo J, Wu T, Anderson J, Kane BF, Johnson DG, Gorelick RJ, Henderson LE, Levin JG: Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J Virol. 2000, 74 (19): 8980-8.
Yu Q, Darlix J-L: The zinc finger of nucleocapsid protein of Friend murine leukemia virus is critical for proviral DNA synthesis in vivo. J Virol. 1996, 70 (9): 5791-8.
Gonsky J, Bacharach E, Goff SP: Identification of residues of the Moloney murine leukemia virus nucleocapsid critical for viral DNA synthesis in vivo. J Virol. 2001, 75 (6): 2616-26.
Darlix JL, Lapadat-Tapolsky M, de Rocquigny H, Roques BP: First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995, 254 (4): 523-37.
Méric C, Goff SP: Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. J Virol. 1989, 63 (4): 1558-68.
Rein A, Harvin DP, Mirro J, Ernst SM, Gorelick RJ: Evidence that a central domain of nucleocapsid protein is required for RNA packaging in murine leukemia virus. J Virol. 1994, 68 (9): 6124-9.
Zhang Y, Qian H, Love Z, Barklis E: Analysis of the assembly function of the human immunodeficiency virus type 1 gag protein nucleocapsid domain. J Virol. 1998, 72 (3): 1782-9.
Feng YX, Copeland TD, Henderson LE, Gorelick RJ, Bosche WJ, Levin JG, Rein A: HIV-1 nucleocapsid protein induces "maturation" of dimeric retroviral RNA in vitro. Proc Natl Acad Sci USA. 1996, 93 (15): 7577-81.
Fossé P, Motté N, Roumier A, Gabus C, Muriaux D, Darlix J-L, Paoletti J: A short autocomplementary sequence plays an essential role in avian sarcoma-leukosis virus RNA dimerization. Biochemistry. 1996, 35 (51): 16601-9.
Darlix J-L, Gabus C, Allain B: Analytical study of avian reticuloendotheliosis virus dimeric RNA generated in vivo and in vitro. J Virol. 1992, 66 (12): 7245-52.
Rein A, Henderson LE, Levin JG: Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem Sci. 1998, 23 (8): 297-301.
Cristofari G, Darlix J-L: The ubiquitous nature of RNA chaperone proteins. Prog Nucleic Acid Res Mol Biol. 2002, 72: 223-68.
Levin JG, Guo J, Rouzina I, Musier-Forsyth K: Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol. 2005, 80: 217-86.
Thomas JA, Gorelick RJ: Nucleocapsid protein function in early infection processes. Virus Res. 2008, 134 (1–2): 39-63.
Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW, Sowder RC, Barsov E, Hood BL, Fisher RJ, Nagashima K, Conrads TP, Veenstra TD, Lifson JD, Ott DE: Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol. 2006, 80 (18): 9039-52.
Roda RH, Balakrishnan M, Hanson MN, Wohrl BM, Le Grice SF, Roques BP, Gorelick RJ, Bambara RA: Role of the Reverse Transcriptase, Nucleocapsid Protein, and Template Structure in the Two-step Transfer Mechanism in Retroviral Recombination. J Biol Chem. 2003, 278 (34): 31536-46.
Lapadat-Tapolsky M, Gabus C, Rau M, Darlix J-L: Possible roles of HIV-1 nucleocapsid protein in the specificity of proviral DNA synthesis and in its variability. J Mol Biol. 1997, 268 (2): 250-60.
Lener D, Tanchou V, Roques BP, Le Grice SF, Darlix J-L: Involvement of HIV-I nucleocapsid protein in the recruitment of reverse transcriptase into nucleoprotein complexes formed in vitro. J Biol Chem. 1998, 273 (50): 33781-6.
Bampi C, Bibillo A, Wendeler M, Divita G, Gorelick RJ, Le Grice SF, Darlix J-L: Nucleotide excision repair and template-independent addition by HIV-1 reverse transcriptase in the presence of nucleocapsid protein. J Biol Chem. 2006, 281 (17): 11736-43.
Grohmann D, Godet J, Mély Y, Darlix JL, Restle T: HIV-1 Nucleocapsid Traps Reverse Transcriptase on Nucleic Acid Substrates. Biochemistry. 2008, 47 (46): 12230-40.
Buckman JS, Bosche WJ, Gorelick RJ: Human immunodeficiency virus type 1 nucleocapsid zn(2+) fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J Virol. 2003, 77 (2): 1469-80.
Basu VP, Song M, Gao L, Rigby ST, Hanson MN, Bambara RA: Strand transfer events during HIV-1 reverse transcription. Virus Res. 2008, 134 (1–2): 19-38.
Fitzon T, Leschonsky B, Bieler K, Paulus C, Schröder J, Wolf H, Wagner R: Proline residues in the HIV-1 NH2-terminal capsid domain: structure determinants for proper core assembly and subsequent steps of early replication. Virology. 2000, 268 (2): 294-307.
Zhang H, Dornadula G, Orenstein J, Pomerantz RJ: Morphologic changes in human immunodeficiency virus type 1 virions secondary to intravirion reverse transcription: evidence indicating that reverse transcription may not take place within the intact viral core. J Hum Virol. 2000, 3 (3): 165-72.
Fassati A, Goff SP: Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J Virol. 2001, 75 (8): 3626-35.
Warrilow D, Harrich D: HIV-1 replication from after cell entry to the nuclear periphery. Curr HIV Res. 2007, 5 (3): 293-9.
Warrilow D, Stenzel D, Harrich D: Isolated HIV-1 core is active for reverse transcription. Retrovirology. 2007, 4: 77-
Arhel NJ, Souquere-Besse S, Munier S, Souque P, Guadagnini S, Rutherford S, Prévost MC, Allen TD, Charneau P: HIV-1 DNA Flap formation promotes uncoating of the pre-integration complex at the nuclear pore. EMBO J. 2007, 26 (12): 3025-37.
McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, Hope TJ: Visualization of the intracellular behavior of HIV in living cells. J Cell Biol. 2002, 159 (3): 441-52.
Brun S, Solignat M, Gay B, Bernard E, Chaloin L, Fenard D, Devaux C, Chazal N, Briant L: VSV-G pseudotyping rescues HIV-1 CA mutations that impair core assembly or stability. Retrovirology. 2008, 5: 57-
Cimarelli A, Darlix JL: Assembling the human immunodeficiency virus type 1. Cell Mol Life Sci. 2002, 59 (7): 1166-84.
Freed EO: HIV-1 gag proteins: diverse functions in the virus life cycle. Virology. 1998, 251 (1): 1-15.
Raposo G, Moore M, Innes D, Leijendekker R, Leigh-Brown A, Benaroch P, Geuze H: Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic. 2002, 3 (10): 718-29.
Basyuk E, Galli T, Mougel M, Blanchard JM, Sitbon M, Bertrand E: Retroviral genomic RNAs are transported to the plasma membrane by endosomal vesicles. Dev Cell. 2003, 5 (1): 161-74.
Grigorov B, Arcanger F, Roingeard P, Darlix JL, Muriaux D: Assembly of infectious HIV-1 in human epithelial and T-lymphoblastic cell lines. J Mol Biol. 2006, 359 (4): 848-62.
Houzet L, Gay B, Morichaud Z, Briant L, Mougel M: Intracellular assembly and budding of the Murine Leukemia Virus in infected cells. Retrovirology. 2006, 3: 12-
Houzet L, Paillart JC, Smagulova F, Maurel S, Morichaud Z, Marquet R, Mougel M: HIV controls the selective packaging of genomic, spliced viral and cellular RNAs into virions through different mechanisms. Nucleic Acids Res. 2007, 35 (8): 2695-704.
Lori F, di Marzo Veronese F, de Vico AL, Lusso P, Reitz MS, Gallo RC: Viral DNA carried by human immunodeficiency virus type 1 virions. J Virol. 1992, 66 (8): 5067-74.
Trono D: Partial reverse transcripts in virions from human immunodeficiency and murine leukemia viruses. J Virol. 1992, 66 (8): 4893-900.
Zhang H, Dornadula G, Pomerantz RJ: Natural endogenous reverse transcription of HIV-1. J Reprod Immunol. 1998, 41 (1–2): 255-60.
Borroto-Esoda K, Boone LR: Equine infectious anemia virus and human immunodeficiency virus DNA synthesis in vitro: characterization of the endogenous reverse transcriptase reaction. J Virol. 1991, 65 (4): 1952-9.
Zhang H, Dornadula G, Pomerantz RJ: Endogenous reverse transcription of human immunodeficiency virus type 1 in physiological microenvironments: an important stage for viral infection of nondividing cells. J Virol. 1996, 70 (5): 2809-24.
Münch J, Rücker E, Ständker L, Adermann K, Goffinet C, Schindler M, Wildum S, Chinnadurai R, Rajan D, Specht A, Giménez-Gallego G, Sánchez PC, Fowler DM, Koulov A, Kelly JW, Mothes W, Grivel JC, Margolis L, Keppler OT, Forssmann WG, Kirchhoff F: Semen-derived amyloid fibrils drastically enhance HIV infection. Cell. 2007, 131 (6): 1059-71.
Roan NR, Münch J, Arhel N, Mothes W, Neidleman J, Kobayashi A, Smith-McCune K, Kirchhoff F, Greene WC: The cationic properties of SEVI underlie its ability to enhance human immunodeficiency virus infection. J Virol. 2009, 83 (1): 73-80.
Tanchou V, Decimo D, Péchoux C, Lener D, Rogemond V, Berthoux L, Ottmann M, Darlix JL: Role of the N-terminal zinc finger of human immunodeficiency virus type 1 nucleocapsid protein in virus structure and replication. J Virol. 1998, 72 (5): 4442-7.
Darlix JL, Garrido JL, Morellet N, Mély Y, de Rocquigny H: Properties, functions, and drug targeting of the multifunctional nucleocapsid protein of the human immunodeficiency virus. Adv Pharmacol. 2007, 55: 299-346.
Houzet L, Morichaud Z, Mougel M: Fully-spliced HIV-1 RNAs are reverse transcribed with similar efficiencies as the genomic RNA in virions and cells, but more efficiently in AZT-treated cells. Retrovirology. 2007, 4: 30-
Houzet L, Morichaud Z, Didierlaurent L, Muriaux D, Darlix JL, Mougel M: Nucleocapsid mutations turn HIV-1 into a DNA-containing virus. Nucleic Acids Res. 2008, 36 (7): 2311-93.
Didierlaurent L, Houzet L, Morichaud Z, Darlix JL, Mougel M: The conserved N-terminal basic residues and zinc-finger motifs of HIV-1 nucleocapsid restrict the viral cDNA synthesis during virus formation and maturation. Nucleic Acids Res. 2008, 36 (14): 4745-53.
Thomas JA, Bosche WJ, Shatzer TL, Johnson DG, Gorelick RJ: Mutations in human immunodeficiency virus type 1 nucleocapsid protein zinc fingers cause premature reverse transcription. J Virol. 2008, 82 (19): 9318-28.
Grigorov B, Décimo D, Smagulova F, Péchoux C, Mougel M, Muriaux D, Darlix JL: Intracellular HIV-1 Gag localization is impaired by mutations in the nucleocapsid zinc fingers. Retrovirology. 2007, 4: 54-
Beltz H, Clauss C, Piémont E, Ficheux D, Gorelick RJ, Roques B, Gabus C, Darlix JL, de Rocquigny H, Mély Y: Structural determinants of HIV-1 nucleocapsid protein for cTAR DNA binding and destabilization, and correlation with inhibition of self-primed DNA synthesis. J Mol Biol. 2005, 348 (5): 1113-26.
Bampi C, Jacquenet S, Lener D, Décimo D, Darlix JL: The chaperoning and assistance roles of the HIV-1 nucleocapsid protein in proviral DNA synthesis and maintenance. Curr HIV Res. 2004, 2 (1): 79-92.
De Rocquigny H, Gabus C, Vincent A, Fournié-Zaluski MC, Roques B, Darlix JL: Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc Natl Acad Sci USA. 1992, 89 (14): 6472-6.
Carteau S, Batson SC, Poljak L, Mouscadet JF, de Rocquigny H, Darlix JL, Roques BP, Käs E, Auclair C: Human immunodeficiency virus type 1 nucleocapsid protein specifically stimulates Mg2+-dependent DNA integration in vitro. J Virol. 1997, 71 (8): 6225-9.
Carteau S, Gorelick RJ, Bushman FD: Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J Virol. 1999, 73 (8): 6670-9.
Yu Q, Ottmann M, Pechoux C, Le Grice S, Darlix JL: Mutations in the primer grip of human immunodeficiency virus type 1 reverse transcriptase impair proviral DNA synthesis and virion maturation. J Virol. 1998, 72 (9): 7676-80.
Williams MC, Rouzina I, Wenner JR, Gorelick RJ, Musier-Forsyth K, Bloomfield VA: Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc Natl Acad Sci USA. 2001, 98 (11): 6121-6.
de Rocquigny H, Shvadchak V, Avilov S, Dong CZ, Dietrich U, Darlix JL, Mély Y: Targeting the viral nucleocapsid protein in anti-HIV-1 therapy. Mini Rev Med Chem. 2008, 8 (1): 24-35.
Yu SF, Baldwin DN, Gwynn SR, Yendapalli S, Linial ML: Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science. 1996, 271 (5255): 1579-82.
Moebes A, Enssle J, Bieniasz PD, Heinkelein M, Lindemann D, Bock M, McClure MO, Rethwilm A: Human foamy virus reverse transcription that occurs late in the viral replication cycle. J Virol. 1997, 71 (10): 7305-11.
Yu SF, Sullivan MD, Linial ML: Evidence that the human foamy virus genome is DNA. J Virol. 1999, 73 (2): 1565-72.
Gabus C, Ivanyi-Nagy R, Depollier J, Bucheton A, Pelisson A, Darlix JL: Characterization of a nucleocapsid-like region and of two distinct primer tRNALys,2 binding sites in the endogenous retrovirus Gypsy. Nucleic Acids Res. 2006, 34 (20): 5764-77.
Volff JN, Brosius J: Modern genomes with retro-look: retrotransposed elements, retroposition and the origin of new genes. Genome Dyn. 2007, 3: 175-90.
Goodier JL, Kazazian HH: Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008, 135 (1): 23-35.
Dolberg D, Fan H: Further characterization of virus-like 30S (VL30) RNA of mice: initiation of reverse transcription and intracellular synthesis. J Gen Virol. 1981, 54 (Pt 2): 281-91.
Ribet D, Harper F, Esnault C, Pierron G, Heidmann T: The GLN family of murine endogenous retroviruses contains an element competent for infectious viral particle formation. J Virol. 2008, 82 (9): 4413-9.
Lee YN, Bieniasz PD: Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 2007, 3 (1): e10-
Dewannieux M, Harper F, Richaud A, Letzelter C, Ribet D, Pierron G: Heidmann T3 Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 2006, 16 (12): 1548-56.
Beauregard A, Curcio MJ, Belfort M: The Take and Give Between Retrotransposable Elements and their Hosts. Annu Rev Genet. 2008, 42: 587-617.
Contreras-Galindo R, Kaplan MH, Leissner P, Verjat T, Ferlenghi I, Bagnoli F, Giusti F, Dosik MH, Hayes DF, Gitlin SD, Markovitz DM: Human endogenous retrovirus K (HML-2) elements in the plasma of people with lymphoma and breast cancer. J Virol. 2008, 82 (19): 9329-36.
Ejtehadi HD, Freimanis GL, Ali HA, Bowman S, Alavi A, Axford J, Callaghan R, Nelson PN: The potential role of human endogenous retrovirus K10 in the pathogenesis of rheumatoid arthritis: a preliminary study. Ann Rheum Dis. 2006, 65 (5): 612-6.
Molès JP, Tesniere A, Guilhou JJ: A new endogenous retroviral sequence is expressed in skin of patients with psoriasis. Br J Dermatol. 2005, 153 (1): 83-9.
Han Y, Wang X, Dang Y, Zheng YH: APOBEC3G and APOBEC3F require an endogenous cofactor to block HIV-1 replication. PLoS Pathog. 2008, 4 (7): e1000095-
Goila-Gaur R, Strebel K: HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology. 2008, 5: 51-
Geuking MB, Weber J, Dewannieux M, Gorelik E, Heidmann T, Hengartner H, Zinkernagel RM, Hangartner L: Recombination of retrotransposon and exogenous RNA virus results in nonretroviral cDNA integration. Science. 2009, 323 (5912): 393-6.
Acknowledgements
The work is supported by ANRS, CNRS, INSERM and Sidaction (France), and by the NIH (USA). Thanks are due to C. Darlix, L. Didierlaurent, Z. Morichaud and D. Muriaux for their contribution to the new results on HIV-1 NC mutants, and to M. Rau (UK) for manuscript corrections.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MM, LH and JLD equally participated in writing the intellectual content and drafting the manuscript. All authors read and approved the manuscript in its final form.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Mougel, M., Houzet, L. & Darlix, JL. When is it time for reverse transcription to start and go?. Retrovirology 6, 24 (2009). https://doi.org/10.1186/1742-4690-6-24
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1742-4690-6-24