Abstract
Although highly penetrant alleles of BRCA1 and BRCA2 have been shown to predispose to breast cancer, the majority of breast cancer cases are assumed to result from the presence of low-moderate penetrant alleles and environmental carcinogens. Non-synonymous single nucleotide polymorphisms (nsSNPs) are hypothesised to contribute to disease susceptibility and approximately 30 per cent of them are predicted to have a biological significance. In this study, we have applied a bioinformatics-based strategy to identify breast cancer-related nsSNPs from 981 carcinogenesis-related genes expressed in breast tissue. Our results revealed a total of 367 validated nsSNPs, 109 (29.7 per cent) of which are predicted to affect the protein function (functional nsSNPs), suggesting that these nsSNPs are likely to influence the development and homeostasis of breast tissue and hence contribute to breast cancer susceptibility. Sixty-seven of the functional nsSNPs presented as commonly occurring nsSNPs (minor allele frequencies ≥ 5 per cent), representing excellent candidates for breast cancer susceptibility. Additionally, a non-uniform distribution of the common functional nsSNPs among different human populations was observed: 15 nsSNPs were reported to be present in all populations analysed, whereas another set of 15 nsSNPs was specific to particular population(s). We propose that the nsSNPs analysed in this study constitute a unique resource of potential genetic factors for breast cancer susceptibility. Furthermore, the variations in functional nsSNP allele frequencies across major population backgrounds may point to the potential variability of the molecular basis of breast cancer predisposition and treatment response among different human populations.
Similar content being viewed by others
Introduction
Mutations of BRCA1[1] and BRCA2[2] confer high breast cancer risk to the carriers. Such highly penetrant mutations are only responsible for a small fraction (~5-10 per cent) of all breast cancer cases,[3, 4] however, suggesting the presence of other, yet to be identified, mutations in other breast cancer predisposition genes [5–7]. Mutations in a number of genes, such as p53,[8]ATM[6] and Chek2,[9] have also been shown to contribute to breast cancer risk in a very small fraction of breast cancer cases. So far, no other high-penetrant breast cancer susceptibility gene has been identified; however, genetic variations including single nucleotide polymorphisms (SNPs) have been hypothesised to act as low-moderate penetrant alleles and contribute to breast cancer, as well as other complex diseases [7, 10–12].
Variations in protein sequence and function are mainly due to the non-synonymous form of SNPs (nsSNPs). The fraction of nsSNPs in the genome is relatively low (~10 per cent of all coding SNPs)[13] compared with other types, but they are more likely to alter the structure, function and interaction of the proteins, and thus constitute a set of candidate genetic factors associated with disease predisposition [14, 15]. Approximately 30 per cent of the nsSNPs are predicted to have biological consequences [16–18]. Several nsSNPs from the proteins acting in a variety of cellular pathways--such as apoptosis,[19] oxidative stress[20] and signal transduction[21]--have already been reported to be associated with an increased/decreased risk of breast cancer.
Several studies have described cancer-relevant nsSNPs;[22–25] however, to our knowledge they have not been studied in the context of expression of genes in a particular tissue. Clearly, in order for genes to be linked to a disease of a tissue, their protein products should somehow influence that particular tissue, either as exogenous proteins (such as hormones) or endogenous proteins (such as the proteins expressed in that tissue) [26, 27]. In this study, we have applied a bioinformatics-based strategy and identified potentially functional nsSNPs from endogenous carcinogenesis-related proteins expressed in breast tissue.
Methods
Genes
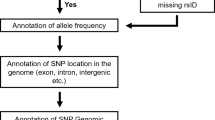
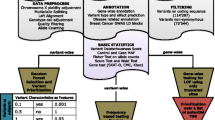
The Ensembl transcript identifiers (http://www.ensembl.org/)[28] of the genes expressed in breast tissue were retrieved from the TissueInfo database (db) (http://icb.med.cornell.edu/services/tissueinfo/query) [29]. The list of carcinogenesis-related genes from 18 different categories ('DNA adduct', 'DNA damage', 'DNA replication', 'angiogenesis', 'apoptosis', 'behavior', 'cell cycle', 'cell signaling', 'development', 'gene regulation', 'transcription', 'immunology', 'metabolism', 'metastasis', 'pharmacology', 'signal transduction', 'tumor suppressors/oncogenes' and 'miscellaneous') was retrieved from the National Cancer Institute's Cancer Genome Anatomy Project Genetic Annotation Initiative ([CGAP-GAI] website [http://lpgws.nci.nih.gov/html-cgap/cgl/]) [30]. The genes retrieved from the TissueInfo and the CGAP-GAI resources were then cross-referenced with each other to identify the group of carcinogenesis-related genes that are expressed in breast tissue.
nsSNPs
The nsSNPs from the group of carcinogenesis-related genes expressed in breast tissue were retrieved from dbSNP build 120 (http://www.ncbi.nlm.nih.gov/SNP/) [31]. Only the nsSNPs detected in ≥ 2 chromosomes in a sample panel of ≥ 40 chromosomes were included in this study (validated nsSNPs). Seventeen nsSNPs were found in both less and more than 5 per cent of the chromosomes analysed in different sample sets; for simplicity, we have classified such nsSNPs within the nsSNP set with ≥ 5 per cent minor allele frequencies throughout this paper.
PolyPhen analysis
The PolyPhen predictions[18] were retrieved from a pre-computed dbSNP-PolyPhen resource. All PolyPhen predictions were based on either alignment of at least five similar proteins (for a more reliable prediction) or structural parameters.
Results
The results obtained in this study are summarised in Table 1 and constitute only the validated nsSNPs with a reliable prediction made by the PolyPhen prediction tool (see Methods). A total of 367 nsSNPs from 189 carcinogenesis-related genes expressed in breast tissue are presented. A total of 109 nsSNPs (28.4 per cent) from 75 genes were predicted potentially to affect the protein function (functional nsSNPs). Additionally, 61.5 per cent (n = 67) of the potentially functional nsSNPs represented commonly occurring nsSNPs in the population (≥ 5 per cent minor allele frequency; Table 2). In this paper, we mainly discuss the commonly occurring functional nsSNPs; however, the list of rarely occurring functional nsSNPs can also be found under the supplementary table (http://www.ozceliklab.com/Breast_rare_nsSNPs/).
A fraction of protein products of genes bearing commonly occurring functional nsSNPs were found to be involved in one or more carcinogenesis-related biological pathways compiled by the CGAP-GAI[30] (Table 2). Such nsSNPs were mostly found in the proteins from DNA repair (three genes, four nsSNPs); metastasis (four genes, four nsSNPs); angiogenesis (seven genes, eight nsSNPs); pharmacology (seven genes, ten nsSNPs); and immunology (38 genes, 51 nsSNPs).
We have also analysed the distribution of the commonly occurring functional nsSNPs across human populations. For simplicity, we have categorised the frequency information obtained from different dbSNP entries into three major groups: African (African and African-American), Caucasian (Caucasian and European) and Asian (Chinese and East Asian) populations. Minor allele frequencies for nsSNPs were available for at least three different human populations for 30 out of 67 commonly occurring functional nsSNPs (Table 3). Fifteen nsSNPs were found in all populations analysed (n ≥ 3). In the case of the remaining 15 nsSNPs, five were found exclusively in one population (ADM-S50R and MMP9-N127K in African; ALDH2-E504K and MNDA-H357Y in Asian; MC1R-R151C in Caucasian). Additionally, three nsSNPs were found in Caucasian, Asian or Hispanic samples, but not in the African samples (CHGA-G382S, CYP1B1-N453S and CYP2C9-R144C). Moreover, in the case of five nsSNPs, the major and the minor alleles were different among the populations analysed (ADBR2-G16R, CDH12-V68M, ERBB2-P1170A, PGM3-D466N and SLC1A5-P17A).
Discussion
A portion of SNPs is considered to contribute to complex disease development [7, 10–12]. SNPs in or around the candidate genes might be directly linked to a disease; however, not all SNPs are supposed to affect gene expression and function, so selection of those with potential effects is keenly debated [32]. Several studies have developed tools and/or systematically analysed nsSNPs to identify those that affect gene function based on evolutionary conservation or structural parameters [16–18, 33]. PolyPhen[18] is one such web-based tool utilised to select the nsSNPs that are likely to affect protein function. In short, the PolyPhen predictions are based on protein alignments, structural parameters or sequence annotations. The sensitivity of PolyPhen has been reported to be approximately 82 per cent [18].
In this study, we hypothesised that the systematic analysis of candidate genes that are expressed in the affected tissue is likely to improve and enrich the identification of disease-susceptibility alleles. Accordingly, using a bioinformatics-based strategy, we identified the functional nsSNPs from a large number of genes related to the carcinogenesis-related pathways (DNA repair, cell cycle, signal transduction, etc), which are expressed in breast tissue. We propose that these potentially functional nsSNPs can result in abnormalities at the protein level, which are likely to affect the development, metabolism and homeostasis of the breast tissue, and thus can contribute to breast cancer susceptibility.
The genes with functional nsSNPs identified in this study were from a variety of carcinogenesis-related cellular pathways. According to this information, possible biological roles for these nsSNPs may be suggested. For example, nsSNPs from angiogenesis- and metastasis-related proteins may have roles in tumour growth and the development of metastatic tumours [34, 35]. Additionally, DNA repair nsSNPs may lead to the accumulation of somatic mutations and thus can participate in cancer initiation and promotion [34–36]. Furthermore, together with the DNA repair nsSNPs, the nsSNPs from the pharmacology genes may also be good candidates for the studies targeting the efficacy, differential response and adverse effect of chemo-/radiotherapy in breast cancer [37–39]. The majority of the nsSNPs were from the genes related to immunological responses (74.6 per cent), which can both suppress and promote tumorigenesis [34]. It is likely that the larger number of the functional nsSNPs in immune system-related genes is a reflection of the large number of immunology genes in the breast tissue-expressed gene set (60 per cent).
A considerable number of genes with functional nsSNPs have been previously linked to breast cancer aetiology: ADM,[40]ADRB2,[41]APOE,[42]CHGA,[43]CSF1,[44]CYP1B1,[45]DAG1,[46]ENG,[47]EPHX1,[48]ERBB2,[49]F2R,[50]MMP9,[51]MUC4,[52]NFATC1,[53]NOTCH4,[54]PLAU,[55]PLAUR,[55]PTGS2[56] and VCAM1 [57]. Therefore, we propose that the nsSNPs in Table 2 are excellent candidates as genetic factors involved in breast cancer initiation, promotion or progression. Additionally, some of these nsSNPs may be critical for breast cancer treatment outcome.
When the distribution of the commonly occurring functional nsSNPs was analysed, differences in the major alleles and the allele frequencies across human populations were observed. For example, 15 commonly occurring nsSNPs were found in all populations, whereas another set of 15 nsSNPs was specific to particular population(s). These differences might be reflections of either the age of the allele, founder effects or the dissimilar selective pressures acting on different populations [58, 59]. Most importantly, the data also indicate that a common nsSNP with a potential biological consequence in our set was equally likely to be either prevalent across different human populations or limited to some populations. Clearly, the latter prompted us to conclude that the population-specific functional nsSNPs may contribute to the genetic predisposition in individuals with a specific background. In this regard, this conclusion is consistent with previous studies in which genetic variations with significantly different allelic frequencies among populations were found to be associated with specific disease or differential drug responses [60–65]. This information may be particularly helpful to researchers in determining which nsSNPs may be relevant to utilise in specific population-based studies. In addition, although further analyses are required, it is tempting to speculate that these nsSNPs may be a part of the potential variability of the molecular basis of breast cancer predisposition and drug response among different human populations.
Data integration from several databases forms the basis of our strategy to determine functional SNPs of breast tissue-expressed genes. The quality and the quantity of the genomic data within individual databases influence the comprehensiveness of the combined data. The functional SNP list presented in this study is a result of data integration from three databases -- namely, TissueInfo,[29] Ensembl,[28] and dbSNP [31]. The non-matching data fields (eg transcript identifiers) between TissueInfo, Ensembl and dbSNP have been the main source of missing data. For example, although BRCA1 was known to have a potentially functional SNP (predicted previously), this information has not been captured because of non-matching transcript identifier information for BRCA1 in the databases. Thus, incompatibility of data in different databases has been a rate-limiting factor for the bioinformatics-based strategies presented here. The improvement of the quality and the quantity of genomic data in the databases will prove beneficial for researching complex questions. Also, the genes presented in this paper are based on the expressed sequence tag information, which may lead to an under-representation of rarely expressed genes [29, 66]. Data integration using other tissue expression databases is likely to enrich the quality of the data produced. Nevertheless, although it is possible that the SNPs presented here may not represent the most comprehensive list, the SNPs identified using the proposed strategy represent a valuable resource for studying the genetic predisposition to breast cancer.
Conclusion
In conclusion, we have designed a novel strategy to identify potentially functional variants of cancer-related genes expressed in breast tissue. Our results demonstrated the presence of 109 nsSNPs with a potential biological consequence, 67 of which were frequent in human populations. We propose that, together with other genetic and environmental factors, these nsSNPs may be involved in breast cancer initiation and progression; thus, these nsSNPs represent the premium candidates as genetic variations of breast cancer predisposition. We also suggest that a considerable fraction of the nsSNPs may, in fact, be population-specific genetic variations.
References
Miki Y, Swensen J, Shattuck-Eidens D, et al: A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994, 266: 66-71. 10.1126/science.7545954.
Wooster R, Bignell G, Lancaster J, et al: Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995, 378: 789-792. 10.1038/378789a0.
Hofmann W, Schlag PM: BRCA1 and BRCA2 -- Breast cancer susceptibility genes. J Cancer Res Clin Oncol. 2000, 126: 487-496. 10.1007/s004320000140.
Hodgson SV, Morrison PJ, Irving M: Breast cancer genetics: Unsolved questions and open perspectives in an expanding clinical practice. Am J Med Genet C Semin Med Genet. 2004, 129: 56-64.
Dong C, Hemminki K: Modification of cancer risks in offspring by sibling and parental cancers from 2,112,616 nuclear families. Int J Cancer. 2001, 92: 144-150. 10.1002/1097-0215(200102)9999:9999<::AID-IJC1147>3.0.CO;2-C.
Chenevix-Trench G, Spurdle AB, Gatei M, et al: Dominant negative ATM mutations in breast cancer families. J Natl Cancer Inst. 2002, 94: 205-215. 10.1093/jnci/94.3.205.
Ponder BA: Cancer genetics. Nature. 2001, 411: 336-341. 10.1038/35077207.
Malkin D, Li FP, Strong LC, et al: Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990, 250: 1233-1238. 10.1126/science.1978757.
Meijers-Heijboer H, van den Ouweland A, Klijn J, et al: Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002, 31: 55-59. 10.1038/ng879.
Risch N, Merikangas K: The future of genetic studies of complex human diseases. Science. 1996, 273: 1516-1517. 10.1126/science.273.5281.1516.
Collins A, Lonjou C, Morton NE: Genetic epidemiology of single-nucleotide polymorphisms. Proc Natl Acad Sci USA. 1999, 96: 15173-15177. 10.1073/pnas.96.26.15173.
Houlston RS, Peto J: The search for low-penetrance cancer susceptibility alleles. Oncogene. 2004, 23: 6471-6476. 10.1038/sj.onc.1207951.
Reumers J, Schymkowitz J, Ferkinghoff-Borg J, et al: SNPeffect: A database mapping molecular phenotypic effects of human non-synonymous coding SNPs. Nucleic Acids Res. 2005, D527-D532. 33 Database
Chanock S: Candidate genes and single nucleotide polymorphisms (SNPs) in the study of human disease. Dis Markers. 2001, 17: 89-98.
Pharoah PD, Dunning AM, Ponder BA, Easton DF: Association studies for finding cancer-susceptibility genetic variants. Nat Rev Cancer. 2004, 4: 850-860. 10.1038/nrc1476.
Wang Z, Moult J: SNPs, protein structure, and disease. Hum Mutat. 2001, 17: 263-270. 10.1002/humu.22.
Ng PC, Henikoff S: Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002, 12: 436-446. 10.1101/gr.212802.
Ramensky V, Bork P, Sunyaev S: Human non-synonymous SNPs: Server and survey. Nucleic Acids Res. 2002, 30: 3894-3900. 10.1093/nar/gkf493.
MacPherson G, Healey CS, Teare MD, et al: Association of a common variant of the CASP8 gene with reduced risk of breast cancer. J Natl Cancer Inst. 2004, 96: 1866-1869. 10.1093/jnci/dji001.
Menzel HJ, Sarmanova J, Soucek P, et al: Association of NQO1 polymorphism with spontaneous breast cancer in two independent populations. Br J Cancer. 2004, 90: 1989-1994. 10.1038/sj.bjc.6601779.
Rutter JL, Chatterjee N, Wacholder S, Struewing J: The HER2 I655V polymorphism and breast cancer risk in Ashkenazim. Epidemiology. 2003, 14: 694-700. 10.1097/01.ede.0000083227.74669.7b.
Livingston RJ, von Niederhausern A, Jegga AG, et al: Pattern of sequence variation across 213 environmental response genes. Genome Res. 2004, 14: 1821-1831. 10.1101/gr.2730004.
Savas S, Kim DY, Ahmad MF, et al: Identifying functional genetic variants in DNA repair pathway using protein conservation analysis. Cancer Epidemiol Biomarkers Prev. 2004, 13: 801-807.
Xi T, Jones IM, Mohrenweiser HW: Many amino acid substitution variants identified in DNA repair genes during human population screenings are predicted to impact protein function. Genomics. 2004, 83: 970-979. 10.1016/j.ygeno.2003.12.016.
Savas S, Ahmad MF, Shariff M, et al: Candidate nsSNPs that can affect the functions and interactions of cell cycle proteins. Proteins. 2005, 58: 697-705.
Ben-Shlomo I, Vitt UA, Hsueh AJ: Perspective: The ovarian kaleidoscope database-II. Functional genomic analysis of an organ-specific database. Endocrinology. 2002, 143: 2041-2044. 10.1210/en.143.6.2041.
Morton CC: Gene discovery in the auditory system using a tissue specific approach. Am J Med Genet A. 2004, 130: 26-28.
Hubbard T, Andrews D, Caccamo M, et al: Ensembl 2005. Nucleic Acids Res. 2005, D447-D453. 33 Database
Skrabanek L, Campagne F: TissueInfo: High-throughput identification of tissue expression profiles and specificity. Nucleic Acids Res. 2001, 29: E102-2. 10.1093/nar/29.21.e102.
Clifford R, Edmonson M, Hu Y, et al: Expression-based genetic/physical maps of single-nucleotide polymorphisms identified by the cancer genome anatomy project. Genome Res. 2000, 10: 1259-1265. 10.1101/gr.10.8.1259.
Sherry ST, Ward MH, Kholodov M, et al: dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29: 308-311. 10.1093/nar/29.1.308.
Daly AK, Day CP: Candidate gene case-control association studies: Advantages and potential pitfalls. Br J Clin Pharmacol. 2001, 52: 489-499. 10.1046/j.0306-5251.2001.01510.x.
Sunyaev S, Ramensky V, Koch I, et al: Prediction of deleterious human alleles. Hum Mol Genet. 2001, 10: 591-597. 10.1093/hmg/10.6.591.
Jakobisiak M, Lasek W, Golab J: Natural mechanisms protecting against cancer. Immunol Lett. 2003, 90: 103-122. 10.1016/j.imlet.2003.08.005.
Kirsch M, Schackert G, Black PM: Metastasis and angiogenesis. Cancer Treat Res. 2004, 117: 285-304. 10.1007/978-1-4419-8871-3_17.
Mohrenweiser HW: Genetic variation and exposure related risk estimation: Will toxicology enter a new era? DNA repair and cancer as a paradigm. Toxicol Pathol. 2004, 32: 136-145.
Andreassen CN, Alsner J, Overgaard M, Overgaard J: Prediction of normal tissue radiosensitivity from polymorphisms in candidate genes. Radiother Oncol. 2003, 69: 127-135. 10.1016/j.radonc.2003.09.010.
Watters JW, McLeod HL: Cancer pharmacogenomics: Current and future applications. Biochim Biophys Acta. 2003, 1603: 99-111.
Sullivan A, Syed N, Gasco M, et al: Polymorphism in wildtype p53 modulates response to chemotherapy in vitro and in vivo. Oncogene. 2004, 23: 3328-3337. 10.1038/sj.onc.1207428.
Oehler MK, Fischer DC, Orlowska-Volk M, et al: Tissue and plasma expression of the angiogenic peptide adrenomedullin in breast cancer. Br J Cancer. 2003, 89: 1927-1933. 10.1038/sj.bjc.6601397.
Cakir Y, Plummer HK, Tithof PK, Schuller HM: Beta-adrenergic and arachidonic acid-mediated growth regulation of human breast cancer cell lines. Int J Oncol. 2002, 21: 153-157.
Zunarelli E, Nicoll JA, Migaldi M, Trentini GP: Apolipoprotein E polymorphism and breast carcinoma: Correlation with cell proliferation indices and clinical outcome. Breast Cancer Res Treat. 2000, 63: 193-198. 10.1023/A:1006464409137.
Pagani A, Papotti M, Hofler H, et al: Chromogranin A and B gene expression in carcinomas of the breast. Correlation of immunocytochemical, immunoblot, and hybridization analyses. Am J Pathol. 1990, 136: 319-327.
Lin EY, Gouon-Evans V, Nguyen AV, Pollard JW: The macrophage growth factor CSF-1 in mammary gland development and tumor progression. J Mammary Gland Biol Neoplasia. 2002, 7: 147-162. 10.1023/A:1020399802795.
Spink DC, Spink BC, Cao JQ, et al: Differential expression of CYP1A1 and CYP1B1 in human breast epithelial cells and breast tumor cells. Carcinogenesis. 1998, 19: 291-298. 10.1093/carcin/19.2.291.
Sgambato A, Migaldi M, Montanari M, et al: Dystroglycan expression is frequently reduced in human breast and colon cancers and is associated with tumor progression. Am J Pathol. 2003, 162: 849-860. 10.1016/S0002-9440(10)63881-3.
Li C, Guo B, Bernabeu C, Kumar S: Angiogenesis in breast cancer: The role of transforming growth factor beta and CD105. Microsc Res Tech. 2001, 52: 437-449. 10.1002/1097-0029(20010215)52:4<437::AID-JEMT1029>3.0.CO;2-G.
Fritz P, Murdter TE, Eichelbaum M, et al: Microsomal epoxide hydrolase expression as a predictor of tamoxifen response in primary breast cancer: A retrospective exploratory study with long-term follow-up. J Clin Oncol. 2001, 19: 3-9.
Zhou BP, Hung MC: Dysregulation of cellular signaling by HER2/neu in breast cancer. Semin Oncol. 2003, 30: 38-48.
Booden MA, Eckert LB, Der CJ, Trejo J: Persistent signaling by dysregulated thrombin receptor trafficking promotes breast carcinoma cell invasion. Mol Cell Biol. 2004, 24: 1990-1999. 10.1128/MCB.24.5.1990-1999.2004.
Lee PP, Hwang JJ, Murphy G, Ip MM: Functional significance of MMP-9 in tumor necrosis factor-induced proliferation and branching morphogenesis of mammary epithelial cells. Endocrinology. 2000, 141: 3764-3773. 10.1210/en.141.10.3764.
Carraway KL, Price-Schiavi SA, Komatsu M, et al: Muc4/sialomucin complex in the mammary gland and breast cancer. J Mammary Gland Biol Neoplasia. 2001, 6: 323-337. 10.1023/A:1011327708973.
Jauliac S, Lopez-Rodriguez C, Shaw LM, et al: The role of NFAT transcription factors in integrin-mediated carcinoma invasion. Nat Cell Biol. 2002, 4: 540-544. 10.1038/ncb816.
Politi K, Feirt N, Kitajewski J: Notch in mammary gland development and breast cancer. Semin Cancer Biol. 2004, 14: 341-347. 10.1016/j.semcancer.2004.04.013.
Sliva D: Signaling pathways responsible for cancer cell invasion as targets for cancer therapy. Curr Cancer Drug Targets. 2004, 4: 327-336. 10.2174/1568009043332961.
Singh B, Lucci A: Role of cyclooxygenase-2 in breast cancer. J Surg Res. 2002, 108: 173-179. 10.1006/jsre.2002.6532.
O'Hanlon DM, Fitzsimons H, Lynch J, et al: Soluble adhesion molecules (E-selectin, ICAM-1 and VCAM-1) in breast carcinoma. Eur J Cancer. 2002, 38: 2252-2257. 10.1016/S0959-8049(02)00218-6.
Cavalli-Sforza LL, Feldman MW: The application of molecular genetic approaches to the study of human evolution. Nat Genet. 2003, 33: 266-275. 10.1038/ng1113.
Fay JC, Wu CI: Sequence divergence, functional constraint, and selection in protein evolution. Annu Rev Genomics Hum Genet. 2003, 4: 213-235. 10.1146/annurev.genom.4.020303.162528.
London SJ, Lehman TA, Taylor JA: Myeloperoxidase genetic polymorphism and lung cancer risk. Cancer Res. 1997, 57: 5001-5003.
Evans DA, McLeod HL, Pritchard S, et al: Interethnic variability in human drug responses. Drug Metab Dispos. 2001, 29: 606-610.
Gibson AW, Edberg JC, Wu J, et al: Novel single nucleotide polymorphisms in the distal IL-10 promoter affect IL-10 production and enhance the risk of systemic lupus erythematosus. J Immunol. 2001, 166: 3915-3922.
Hopper JL: Genetic epidemiology of female breast cancer. Semin Cancer Biol. 2001, 11: 367-374. 10.1006/scbi.2001.0392.
Xu C, Goodz S, Sellers EM, Tyndale RF: CYP2A6 genetic variation and potential consequences. Adv Drug Deliv Rev. 2002, 54: 1245-1256. 10.1016/S0169-409X(02)00065-0.
Shimizu E, Hashimoto K, Iyo M: Ethnic difference of the BDNF 196G/A (val66met) polymorphism frequencies: The possibility to explain ethnic mental traits. Am J Med Genet B Neuropsychiatr Genet. 2004, 126: 122-123.
Wang SM, Rowley JD: A strategy for genome-wide gene analysis: Integrated procedure for gene identification. Proc Natl Acad Sci USA. 1998, 95: 11909-11914. 10.1073/pnas.95.20.11909.
Povey S, Lovering R, Bruford E, et al: The HUGO Gene Nomenclature Committee (HGNC). Hum Genet. 2001, 109: 678-680. 10.1007/s00439-001-0615-0.
Acknowledgements
The authors thank Baris Tuncertan and Mehjabeen Shariff for retrieving the data from the dbSNP and the pre-computed PolyPhen resource and Dr Michelle Cotterchio for critically reading the manuscript. This work was supported by grants (BCTR0100627) from the Susan Komen Breast Cancer Foundation, USA, and the Canadian Breast Cancer Foundation. Sevtap Savas is supported, in part, by a 'CIHR Strategic Training Program Grant -- The Samuel Lunenfeld Research Institute Training Program: Applying Genomics to Human Health' fellowship.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Savas, S., Schmidt, S., Jarjanazi, H. et al. Functional nsSNPs from carcinogenesis-related genes expressed in breast tissue: Potential breast cancer risk alleles and their distribution across human populations. Hum Genomics 2, 287 (2006). https://doi.org/10.1186/1479-7364-2-5-287
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1479-7364-2-5-287