Abstract
Background
The endometrium is a dynamic tissue whose changes are driven by the ovarian steroidal hormones. Its main function is to provide an adequate substrate for embryo implantation. Using microarray technology, several reports have provided the gene expression patterns of human endometrial tissue during the window of implantation. However it is required that biological connections be made across these genomic datasets to take full advantage of them. The objective of this work was to perform a research synthesis of available gene expression profiles related to acquisition of endometrial receptivity for embryo implantation, in order to gain insights into its molecular basis and regulation.
Methods
Gene expression datasets were intersected to determine a consensus endometrial receptivity transcript list (CERTL). For this cluster of genes we determined their functional annotations using available web-based databases. In addition, promoter sequences were analyzed to identify putative transcription factor binding sites using bioinformatics tools and determined over-represented features.
Results
We found 40 up- and 21 down-regulated transcripts in the CERTL. Those more consistently increased were C4BPA, SPP1, APOD, CD55, CFD, CLDN4, DKK1, ID4, IL15 and MAP3K5 whereas the more consistently decreased were OLFM1, CCNB1, CRABP2, EDN3, FGFR1, MSX1 and MSX2. Functional annotation of CERTL showed it was enriched with transcripts related to the immune response, complement activation and cell cycle regulation. Promoter sequence analysis of genes revealed that DNA binding sites for E47, E2F1 and SREBP1 transcription factors were the most consistently over-represented and in both up- and down-regulated genes during the window of implantation.
Conclusions
Our research synthesis allowed organizing and mining high throughput data to explore endometrial receptivity and focus future research efforts on specific genes and pathways. The discovery of possible new transcription factors orchestrating the CERTL opens new alternatives for understanding gene expression regulation in uterine function.
Similar content being viewed by others
Background
The human endometrium is a complex tissue whose cyclic regulation is mainly driven by the changing pattern of the ovarian steroidal hormones estradiol (E2) and progesterone (P4) [1]. The main function of the endometrium is to provide receptive substrate at the appropriate time for blastocyst implantation. Although it is non-adhesive to embryos throughout most of the menstrual cycle [2] the action of P4 on an E2-primed endometrium induces a certain gene expression profile that is favorable for blastocyst adhesion during a restricted period of time known as the 'window of implantation' [3, 4]. In women, this maternally directed receptive phase appears to be of approximately 5 days' duration, from day 20 to day 24 of a 28-days menstrual cycle [5]. The molecular basis of the window of implantation in human endometrium is beginning to be unrevealed and a number of biochemical markers for uterine receptivity have been proposed [3, 6].
Microarrays analysis, an assay that is used to measure the level of mRNA expression of thousands of genes in a group of cells [7], enables discovery of genes or pathways likely to be involved in a biological process. This approach has been used to broadly characterize the molecular bases of endometrial function in women, by determining the gene expression profiles corresponding to each endometrial phase during the menstrual cycle [8–10]. In addition, it has been used to specifically investigate the acquisition of endometrial receptivity to embryo implantation during spontaneous cycles [11–15]. Since changes in the endometrium toward acquisition of receptivity are mainly driven by progesterone (P4) [16, 17], two strategies have been used for gene discovery during spontaneous menstrual cycles. These are based on the comparison of the endometrial transcriptome under peak P4 circulating levels (days 19-23, window of implantation) compared to the endometrial gene expression profiles obtained under absent (days 8-11, proliferative phase) [11, 12] or low (days 15-17, early secretory phase) [13–15, 18, 19] serum P4.
Although DNA microarrays are a powerful tool for gene discovery, there are several substantial sources of noise in microarray data. Intra- and inter-microarray variations limit the statistical power to discriminate the differentially expressed genes. While validation of microarray data is required to overcome this issue, most reports of endometrial gene expression analysis included validation of only a small number of differentially expressed genes (usually less than 10) by an independent mRNA quantification method (Northern blot, semi-quantitative or quantitative RT-PCR) [20]. Integration and cross-validation of data sets about endometrial gene expression profiles produced by different groups could increase confidence in gene expression results for many more genes than is tractable with classical validation [21, 22] and should provide the up- and down-regulated genes that together orchestrate the acquisition of the receptive phenotype of the endometrium. Such exploration and integration could help researchers to obtain a comprehensive view of existing data and better prioritize experimental efforts.
Transcriptional regulatory mechanisms are crucial for temporal and spatial gene expression. These mechanisms are mediated by a set of transcription factors (TFs), proteins which have the ability to bind to a specific region on the gene (known as motifs or transcription factor binding sites (TFBS)), to regulate transcription. It is thought that co-expression of genes frequently arises from transcriptional co-regulation. As co-regulated genes share some similarities in their regulatory mechanism, possibly at transcriptional level, their promoter regions may contain common motifs that are binding sites for transcription regulators [23]. Given a cluster of endometrial regulated genes with similar expression profiles, the characterization of their regulatory regions is a fundamental step toward understanding the largely unexplored networks of gene regulation in this complex tissue responsible for their coordinated behavior. Computation biology of gene regulation offers several bioinformatic tools developed for the prediction of TFBS within a specific regulatory DNA sequence [24]. Given a set of co-regulated transcripts, in silico predictions of TFBS in their regulatory regions offers a unique opportunity to identify novel components, leading to the formulation of transcriptional regulatory networks hypotheses that can be further tested in the wet laboratory.
The aim of this study was to increase our understanding of endometrial receptivity to embryo implantation, by performing a research synthesis of the publicly available DNA microarray data. The first objective was to determine genes consistently reported in the literature as either up- or down-regulated from pre receptive to the receptive endometrium. The second objective was to identify possible TFs that may mediate the regulation of endometrial gene expression, by analyzing the cis-regulatory sequences of genes sharing a common regulatory behavior.
Methods
Integration and cross-validation of microarrays data
The available data sets comparing endometrial gene expression profiles from the proliferative vs. mid secretory phase [11, 12] and from early secretory vs. mid secretory phase [9, 13–15, 19] were analyzed (Table 1). The UniGene key identifier (cluster ID) for each differential expressed transcript was obtained from the SOURCE [25], NetAffx [26] and UniGene [27] databases. Each UniGene entry is a set of transcript sequences that appear to come from the same transcription locus (gene or expressed pseudogene) and was used for cross-referencing transcripts amongst databases. The information from each database was imported into Microsoft Access® software and used as a relational database to determine transcripts that show consistent differential expression under similar experimental conditions. Those having a similar transcriptional response (up- or down-regulation) in at least 4 reports for increased and 3 for decreased transcripts were considered biologically relevant and included in a list we have designated the 'consensus endometrial receptivity transcript list' (CERTL). The difference in threshold for considering down-regulated transcripts is because the study from Haouzi et al 2009 [18] does not disclose the decreased transcripts.
Functional clustering
Those up- and down-regulated genes from the CERTL were submitted to web-based databases for functional annotation analysis in order to gain an in-depth understanding of the biological themes in the CERTL. DAVID (Database for Annotation, Visualization and Integrated Discovery) [28] and GATHER (Gene Annotation Tool to Help Explain Relationships) [29] webtools were used for this purpose. Both services extract the biological meaning of submitted genes by retrieving their functional annotations from the Kyoto Encyclopedia of Genes and Genomes (KEGG) [30], Biocarta pathways [31] and Gene Ontology (GO) [32] databases.
TFBS detection in promoter regions of genes associated to endometrial receptivity
We firstly examined the promoter region of our genes of interest defined as the region proximal to the transcription-start site of genes transcribed by RNA polymerase II. For a systematic search for potential TFBS, we used the following approaches and platforms to increase the power of our results:
MotifScanner. We used the stand-alone version of Motifscanner [33] that searches for potential TFBSs in a set of sequences using all the TRANSFAC vertebrate position-weigh matrices (PWMs) [34]. The information of TFBS obtained from MotifScanner was sent to the software TOUCAN [23] for determination of PWMs that were significantly over-represented.
Over-represented Transcription Factor Binding Site Prediction Tool (OTFBS). This web-tool [35, 36] searches for potential TFBSs based on the TRANSFAC PWMs using the MatInspector algorithm [37] and determines over-represented motifs in regulatory sequences.
The Transcription Element Listening System (TELiS). The TELiS database [38, 39] uses the TRANSFAC and JASPAR [40] PWMs in order to detect potential TFBS. It uses the MatInspector algorithm through the Java application PromoterScan [38] and identifies over-represented motifs.
GATHER. This database [29] searches for potential TFBS using TRANSFAC 8.2 PWMs [41, 42] and identifies statically over-represented TFBS.
Results
Identification of genes associated to endometrial receptivity
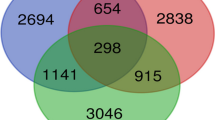
We intersected the lists of regulated genes reported in studies using microarrays analysis of endometrial receptivity for determining those consistently regulated across different reports. As expected the number of coincident genes was small, considering the number of genes comprising each list. We identified 40 up-regulated genes in at least four of seven reports (Table 2) and 21 down-regulated genes present in at least three of six studies considered (Tables 3), collectively denominated CERTL. The most consistent up-regulated genes were C4BPA, SPP1, APOD, CD55, CFD, CLDN4, DKK1, ID4, IL15 and MAP3K5; whereas OLFM1, CCNB1, CRABP2, EDN3, FGFR1, MSX1 and MSX2 were the most consistently down-regulated in endometrial tissue for the acquisition of receptivity to embryo implantation.
Functional associations of transcripts from CERTL
To gain further understanding of the potential functional roles of regulated transcripts present in CERTL we obtained the functional annotations from each gene and determined the enriched processes from two different web-based tools. The up-regulated transcript list was consistently enriched with transcripts related to the immune response and complement activation whereas the down-regulated transcript list was enriched with transcripts related to cell cycle regulation (Tables 4 and 5).
Identification of consensus sequences for TFBS sites of CERTL
We hypothesized that genes showing a common regulatory behavior may also share common regulatory mechanisms such as TFBSs in their respective promoter regions. To identify these possible common regulatory patterns that should be over-represented in the CERTL, we took advantage of several publicly available bioinformatics tools. The potential TFBS were detected in a first step, and then those statistically over-represented in our endometrial gene cluster were determined. The results are listed in Table 6 for up- and down-regulated transcripts respectively. Interestingly, DNA binding sites for E47, Sterol Regulatory Element Binding Protein 1 (SREBP1) and E2F1 were the most consistently over-represented and present in both up- and down-regulated transcripts. The number of increased genes with predicted TFBS for E2F1, SREBP1 and E47 was at least 20, 13 and 7 respectively in a total of 40. Of 21 decreased genes the number of transcripts with predicted TFBS was at least 14, 2 and 3 respectively. Other TFs over-represented were MEF2, FREAC2 and ARNT.
Discussion
Scientific knowledge of how endometrial receptivity is regulated is fundamental for the understanding of the mechanisms that govern embryonic implantation. The availability of public datasets related to global endometrial gene regulation during the acquisition of the receptive phenotype, provides a tool for the analysis of regulation of gene expression using bioinformatics tools. Using DNA microarrays analysis, several approaches have been used for determining the genes of uterine receptivity assessing the endometrium in different physiological [9, 11–15, 18, 43], pathological [44–48] or intervened conditions [49, 50]. We here analyzed seven reports of endometrial gene expression profiling during spontaneous cycles: Carson et al. [13], Kao et al. [11], Borthwick et al. [12], Riesewijk et al. [14], Mirkin et al. [15], Talbi et al. [9] and Haouzi et al. [18]. Since the number of endometrial samples analyzed in each of these studies was limited, the question arises as to whether the groups investigated were representative of the population. This is a major concern for any statistical analysis. Therefore we considered all studies together in a research synthesis to provide a larger sample size thus consolidating the selection of actual regulated transcripts in the endometrium. A first step was to associate probes and available annotations in the reports that belong to the same UniGene cluster (i.e. with same UniGene ID), and then proceed to further comparisons to identify common transcripts that are similarly regulated during the window of implantation. Previous partial analyses [15, 43, 51–53] found very few transcripts to be consistently regulated. In our study we found 61 transcripts regulated in the same direction in the endometrium during the window of implantation; 40 were up-regulated in at least 4 of 7 studies and 21 were down-regulated in at least 3 of 6 reports analyzed.
The relatively small number of consistently regulated transcripts identified could be explained by the differences in the study design, number of samples included and the methodology used for data analysis. However, other factors should be considered when interpreting gene expression analyses related to endometrial receptivity. Importantly, the reports included here, all used RNA extracted from whole endometrial biopsies, tissue that comprises a number of different cell types, including epithelial (luminal and glandular), stromal fibroblasts, endothelial cells, vascular smooth muscle cells and lymphoid cells. Hence the endometrial changes induced by E2 and P4 result from the differential response of each cell type to the same hormones. A clear example is the down regulation of the PR during the secretory phase in endometrial epithelial cells but not in the stromal compartment [54]. Microdissection of cell subpopulations (for example, with laser capture [55]) may disclose the actual gene expression profiles of each cell subpopulations within the tissue context. In addition, any biopsy sample may not represent the complete endometrium since microenvironments occur within this tissue. Nevertheless gene expression profiling of endometrial biopsies during the window of implantation is one of the most promising strategies for gene discovery related to uterine receptivity.
The intersection of gene lists performed in the present study showed that most consistently increased transcripts during the window of implantation were C4BPA, SPP1, APOD, CD55, CFD, CLDN4, DKK1, ID4, IL15 and MAP3K5 whereas OLFM1, CCNB1, CRABP2, EDN3, FGFR1, MSX1 and MSX2 were the most consistently decreased. However, correlation of transcript abundance change with changes in the corresponding protein, followed by functional testing of the biological effect of that protein, is necessary to confirm the biological significance of the microarray changes.
The functional annotations of up-regulated genes within the CERTL showed a significant association to the immune response and complement activation. Most of these genes belong to the innate immune system, which is the immunological first line of defense that provides an immediate response through its ability to distinguish between 'infectious non-self' and 'non-infectious self' [56]. Therefore, innate immunity regulation in the endometrium is of fundamental significance for establishing a microenvironment that will provide adequate tolerance to the implanting embryo [57]. Regarding complement system regulatory proteins, their possible roles and expression levels in the endometrium throughout the normal menstrual cycle have been reported [58–62]. Most of these studies show an increase of complement-regulatory molecules during the secretory phase in human endometrium [58, 61, 62] in line with the increased mRNA levels of the complement system molecules C4b-binding protein (C4BP) and adipsin (complement component factor D, CFD) from the CERTL. It is postulated that the complement system might be conferring immunity to the uterine cavity, defending it against bacterial infection. In this sense, C4BP may provide a protective role to the embryo where an increased expression of an inhibitor of complement system activation could reduce the chance of a misdirected complement attack to the embryo (which is considered as a semiallograft). Indeed, C4BPA transcript levels are abnormally decreased in the endometrium during the receptive phase in women with endometriosis [44, 63], implantation failure [46] and unexplained recurrent abortion [64], suggesting it may have a role in embryo implantation. By contrast, adipsin may have a non-complement function in the female reproductive tract as suggested for other complement-molecules [60]. Adipsin is necessary for the production of oviduct-derived embryotrophic factor-3 (ETF-3) [65, 66] which stimulates embryo development [67, 68]. Thus up-regulation of adipsin in human endometrium may assist the embryo during the implantation process as shown for other chemokines in the endometrium [69].
Several down-regulated genes within CERTL are associated with cell cycle regulation, including cyclin B1 (CCNB1) the most consistently down-regulated gene. CCNB1 binds to p34 (cdc2) to form the mitosis-promoting factor during G2 phase [70, 71]. In human secretory phase endometrium, CCNB1 is decreased compared to the proliferative phase [72, 73] supporting the microarray data used to construct the CERTL. Moreover, in endometrial cell cultures, P4 decreases the expression of CCNB1, inhibits cell proliferation and induces apoptosis, suggesting that cyclin B1 may play an important role in proliferation and differentiation of the endometrial tissue under steroidal regulation.
Cellular retinol binding protein-2 (CRABP2) is a cytosolic protein that binds retinoic acid (RA) with high affinity [74]. The CRABP2 transcript has been reported to decrease from the proliferative to the secretory phase in human endometrium [75], which is in line with the microarrays reports used for constructing our CERTL. The physiological effects of RA are mediated by members of two families of nuclear receptors [76, 77] and they all have been detected by immunohistochemistry in human endometrium throughout the phases of the menstrual cycle [78] in epithelial and stromal cells. The fact that CRABP2 decreases in human endometrium at the time of embryo implantation might suggest that RA signaling is required to be silenced, since it shuttles RA to the RA receptors in the cell nucleus [74, 78]. In the mouse uterus, CRABP2 decreases around the time of embryo implantation [79] whereas P4 induces the expression of cyp26a1, the enzyme responsible for RA catabolism in mouse uterine epithelial cells [80, 81]. Knock down of cyp26a1 in mouse uterus decreases embryo implantation rate [82]. In addition, in human secretory endometrium, cyp26a1 mRNA level is ~20 times higher than in the proliferative phase [83]. Since the action of RA is essential for endometrial stromal cell decidualization [79] silencing of RA signaling during the window of implantation might prevent precocious decidualization of stromal cells that could compromise endometrial receptivity.
The cytokine endothelin-3 (EDN3) and fibroblast growth factor receptor-1 (FGFR1) were among the transcripts consistently down-regulated in the endometrium during the window of implantation. There is abundant evidence showing that both endometrial receptivity and blastocyst implantation are regulated by cytokines and growth factors [84]. Immunoreactive pro-endothelin-3 has been described in human endometrium in luminal and glandular epithelia; however cycle-dependent regulation of this molecule is not clear [85]. Its action in the human endometrium is suggested to be in paracrine vasoactive control of the uterine vascular bed [86]. However this cytokine has many other functions such as proliferation and development of several cell types [87–90]. In the mouse oviduct, EDN3 signaling has been associated with the regulation of transcripts related to TGFβ, IL-10, and C/EBP [91]. Its functional role in the human endometrium and the effects of its down-regulation during the window of implantation has yet to be determined. FGFR1 and its ligand FGF-2 have also been described in human endometrium [92–95]. Immunoreactive FGFR1 and its transcript are significantly higher in proliferative that in secretory human endometrium [93, 94] supporting the down-regulation of this transcript included in the CERTL. However, not all studies have reported such endometrial regulation [95]. FGF-2 promotes endometrial stromal proliferation [94, 96] and ovarian steroid hormones modulate its synthesis and function in endometrial cells [96, 97]. The functional relevance of FGFR1 down-regulation in endometrial receptivity remains to be elucidated.
With regard to the TFs present in the CERTL, we found the inhibitor of DNA binding 4 (ID4) up-regulated and MSX-1 and -2 down-regulated. In animal models, uterine MSX-1 and -2 are down-regulated by P4 [98] or during embryo implantation [99–101]. Constant expression of Msx1 in the infertile Lif-/- mice uterus further supports a role for MSX-1down-regulation in endometrial receptivity [100]. ID4 TF is a member of a family of inhibitor of DNA binding proteins (Id) that has been associated with cell proliferation and differentiation [102–105]. Its regulatory effect in human endometrium is unknown. Many other TFs associated with endometrial regulation [106–120] have provided insights into the molecular basis of gene regulation for endometrial function in response to sex steroid hormones. We reasoned that the cluster of regulated genes derived from microarray experiments related to endometrial receptivity (i.e. CERTL) would allow a different strategy for TF discovery, namely comparative promoter analysis. This is based on the hypothesis that genes showing a common regulatory behavior may also share common regulatory mechanisms such as TFBSs in their respective promoter regions. Interestingly, we found that E47, E2F1 and SREBP1 are common TFBSs for up- and down-regulated transcripts from CERTL so it is likely that they orchestrate the changes in transcript profile for endometrial receptivity. None of these three TFs have been described in normal human endometrium in the context of their regulation during the menstrual cycle, in response to steroidal hormones or a regulatory role on uterine function. However, there is no guarantee that the revealed TFBS are indeed functional in the context of regulatory regions, hence biological verification is required.
The E2F1 TF belongs to the E2F family [121] and displays properties of both an oncogene (induction of proliferation) and tumor suppressor (induction of apoptosis) [122, 123]. E47 is a TF that belongs to the class I bHLH proteins, also known as E proteins [124] which form homo- or hetero-dimers and bind to specific DNA sequences [125]. Sterol regulatory element-binding protein 1 (SREBP1) is a membrane-bound TFs that belongs to a family of basic helix-loop-helix-leucine zipper (bHLHLZ) TFs [126]. Upon activation, SREBP1 translocates into the nucleus where it binds to sterol regulatory sites located in the promoter regions of genes involved in cholesterol homeostasis and transport [127, 128] such as the steroidogenic acute regulatory protein (StAR), a key regulator of steroidogenesis [129]. Function of bHLH TFs such as E47 can be blocked by Inhibitor of DNA binding (Id) TFs [130, 131]. In addition, SREBP1 as a member of bHLHLZ family, may also be subjected to regulation by Id proteins [132]. In the CERTL ID4 transcript was up-regulated in the receptive endometrium: as a consequence E47 and SREBP-1 TFs may be less available for binding to DNA in target sequences and direct co-regulated transcripts. Interestingly, the TF E2F1 is involved in the transcriptional control of id4 gene expression [133], supporting our bioinformatics findings of overrepresented TFBSs.
It is well known that P4 is essential for the establishment and maintenance of pregnancy in the women and in this sense the study of its actions in the uterus has been focused on changes in gene expression [134, 135]. Responses to P4 in reproductive tissues occur by the activation of classical nuclear P4 receptors (PRA and B), which upon binding with their ligand, function as TFs regulating gene expression [136]. In addition, many transcriptional actions of P4 require interactions with corepressors and coactivators [137–139]. However, P4 may also act in the uterus through at least two families of nonclassical membrane progestin receptors [140, 141]. Hence the genomic and non-genomic pathways may interact and integrate to ultimately affect endometrial gene expression. Interestingly, two of the endometrial transcripts more consistently up-regulated during the mid-secretory phase, APOD and SPP1, do not display progesterone response elements in their cis-regulatory sequences [12, 15] suggesting that P4 induction is not directly mediated by the ligand-bound PR. Interestingly both APOD and SPP1 genes display TFBS for E2F1 in their upstream regulatory sequences. In breast cancer cells, P4 up-regulates the expression of E2F1 and hence indirectly affects transcription of classic E2F1 target genes [115]. Such regulation of E2F1 induced by progestins has been shown to be multimodal since ligand-bound PR can regulate its transcription directly but also indirectly through other molecules to achieve further progestin-mediated regulation of E2F1 expression [142]. Whether E2F1 along with E47 and SRBP1 are also mediating the P4 transcriptional regulation in the endometrium for acquisition of receptivity has yet to be determined.
Identification of the CERTL and the possible regulatory TFs in the present research synthesis should not be viewed as an end in itself. Their real value increases only as these results move through to biological validation, ranging from the numerical verification of expression levels with alternative techniques, to ascertaining the actual regulatory role of the TFs in the endometrial transcriptional networks. Finally, for several transcripts contained in the CERTL, biological knowledge is completely lacking in relation to endometrial physiology, so extensive research is required to better understand the mechanisms underlying endometrial receptivity.
Conclusion
In conclusion, a CERTL comprised of 61 transcripts consistently regulated in human endometrium during the receptive period for embryo implantation has been identified in this study. These transcripts are mainly involved in immune response, complement activation and cell cycle regulation; suggesting that these biological process are associated with the acquisition of the receptive phenotype. Finally, TFBS for E47, SREBP1 and E2F1 were over-represented in the regulatory region of genes from CERTL, suggesting that they may be mediating the effects of the ovarian steroidal hormones in the endometrial transcriptional regulation. Biological validation of such bioinformatic predictions will shed light on the transcriptional networks associated to uterine receptivity for embryo implantation. Moreover, this knowledge can potentially be applied to improve fertility in infertile patients.
Abbreviations
- bHLH:
-
basic helix-loop-helix transcription factors
- bHLHLZ:
-
basic helix-loop-helix-leucine zipper
- C/EBP:
-
CAAT/enhancer-binding proteins
- C4BP:
-
C4b-binding protein
- CCNB1:
-
cyclin B1
- CERTL:
-
consensus endometrial receptivity transcript list
- CFD:
-
complement component factor D
- CLDN4:
-
claudin-4
- CRABP2:
-
cellular retinol binding protein-2
- DAVID:
-
database for annotation, visualization and integrated discovery
- DNA:
-
deoxyribonucleic acid
- E2:
-
estradiol
- EDN3:
-
endothelin-3
- EDNRB:
-
endothelin receptor type B
- ETF-3:
-
oviduct-derived embryotrophic factor-3
- FGF:
-
fibroblast growth factor
- FGFR1:
-
fibroblast growth factor receptor-1
- GATHER:
-
gene annotation tool to help explain relationships
- GO:
-
gene ontology
- Id:
-
inhibitor of DNA binding proteins
- ID4:
-
inhibitor of DNA binding 4
- IL-10:
-
interleukin-10
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- LH:
-
luteinizing hormone
- LIF:
-
leukemia inhibitory factor
- mRNA:
-
messenger ribonucleic acid
- OTFBS:
-
over-represented transcription factor binding site prediction tool
- P4:
-
progesterone
- RA:
-
retinoic acid
- RT-PCR:
-
reverse transcription-polymerase chain reaction
- SRE:
-
sterol regulatory element
- SREBP1:
-
sterol regulatory element binding protein 1
- TELiS:
-
the transcription element listening system
- TF:
-
transcription factor
- TFBS:
-
transcription factor binding site
- TGFβ:
-
transforming growth factor-β.
References
Wynn RM: Cyclic and gestational changes. 1989, New York: Plenum Publishing Corp
Psychoyos A: Uterine receptivity for nidation. Ann N Y Acad Sci. 1986, 476: 36-42. 10.1111/j.1749-6632.1986.tb20920.x.
Giudice LC: Potential biochemical markers of uterine receptivity. Hum Reprod. 1999, 14 (Suppl 2): 3-16.
Salamonsen LA, Nie G, Dimitriadis E, Robb L, Findlay JK: Genes involved in implantation. Reprod Fertil Dev. 2001, 13 (1): 41-49. 10.1071/RD00046.
Navot D, Scott RT, Droesch K, Veeck LL, Liu HC, Rosenwaks Z: The window of embryo transfer and the efficiency of human conception in vitro. Fertil Steril. 1991, 55 (1): 114-118.
Hoozemans DA, Schats R, Lambalk CB, Homburg R, Hompes PG: Human embryo implantation: current knowledge and clinical implications in assisted reproductive technology. Reprod Biomed Online. 2004, 9 (6): 692-715. 10.1016/S1472-6483(10)61781-6.
Schena M, Shalon D, Davis RW, Brown PO: Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995, 270 (5235): 467-470. 10.1126/science.270.5235.467.
Ponnampalam AP, Weston GC, Trajstman AC, Susil B, Rogers PA: Molecular classification of human endometrial cycle stages by transcriptional profiling. Mol Hum Reprod. 2004, 10 (12): 879-893. 10.1093/molehr/gah121.
Talbi S, Hamilton AE, Vo KC, Tulac S, Overgaard MT, Dosiou C, Le Shay N, Nezhat CN, Kempson R, Lessey BA, et al: Molecular phenotyping of human endometrium distinguishes menstrual cycle phases and underlying biological processes in normo-ovulatory women. Endocrinology. 2006, 147 (3): 1097-1121. 10.1210/en.2005-1076.
Punyadeera C, Dassen H, Klomp J, Dunselman G, Kamps R, Dijcks F, Ederveen A, de Goeij A, Groothuis P: Oestrogen-modulated gene expression in the human endometrium. Cell Mol Life Sci. 2005, 62 (2): 239-250. 10.1007/s00018-004-4435-y.
Kao LC, Tulac S, Lobo S, Imani B, Yang JP, Germeyer A, Osteen K, Taylor RN, Lessey BA, Giudice LC: Global gene profiling in human endometrium during the window of implantation. Endocrinology. 2002, 143 (6): 2119-2138. 10.1210/en.143.6.2119.
Borthwick JM, Charnock-Jones DS, Tom BD, Hull ML, Teirney R, Phillips SC, Smith SK: Determination of the transcript profile of human endometrium. Mol Hum Reprod. 2003, 9 (1): 19-33. 10.1093/molehr/gag004.
Carson DD, Lagow E, Thathiah A, Al-Shami R, Farach-Carson MC, Vernon M, Yuan L, Fritz MA, Lessey B: Changes in gene expression during the early to mid-luteal (receptive phase) transition in human endometrium detected by high-density microarray screening. Mol Hum Reprod. 2002, 8 (9): 871-879. 10.1093/molehr/8.9.871.
Riesewijk A, Martin J, van Os R, Horcajadas JA, Polman J, Pellicer A, Mosselman S, Simon C: Gene expression profiling of human endometrial receptivity on days LH+2 versus LH+7 by microarray technology. Mol Hum Reprod. 2003, 9 (5): 253-264. 10.1093/molehr/gag037.
Mirkin S, Arslan M, Churikov D, Corica A, Diaz JI, Williams S, Bocca S, Oehninger S: In search of candidate genes critically expressed in the human endometrium during the window of implantation. Hum Reprod. 2005, 20 (8): 2104-2117. 10.1093/humrep/dei051.
Conneely OM, Mulac-Jericevic B, DeMayo F, Lydon JP, O'Malley BW: Reproductive functions of progesterone receptors. Recent Prog Horm Res. 2002, 57: 339-355. 10.1210/rp.57.1.339.
Spencer TE, Bazer FW: Biology of progesterone action during pregnancy recognition and maintenance of pregnancy. Front Biosci. 2002, 7: d1879-1898. 10.2741/spencer.
Haouzi D, Assou S, Mahmoud K, Tondeur S, Reme T, Hedon B, De Vos J, Hamamah S: Gene expression profile of human endometrial receptivity: comparison between natural and stimulated cycles for the same patients. Hum Reprod. 2009, 24 (6): 1436-1445. 10.1093/humrep/dep039.
Haouzi D, Mahmoud K, Fourar M, Bendhaou K, Dechaud H, De Vos J, Reme T, Dewailly D, Hamamah S: Identification of new biomarkers of human endometrial receptivity in the natural cycle. Hum Reprod. 2009, 24 (1): 198-205. 10.1093/humrep/den360.
White CA, Salamonsen LA: A guide to issues in microarray analysis: application to endometrial biology. Reproduction. 2005, 130 (1): 1-13. 10.1530/rep.1.00685.
Kemmeren P, van Berkum NL, Vilo J, Bijma T, Donders R, Brazma A, Holstege FC: Protein interaction verification and functional annotation by integrated analysis of genome-scale data. Mol Cell. 2002, 9 (5): 1133-1143. 10.1016/S1097-2765(02)00531-2.
Rhodes DR, Barrette TR, Rubin MA, Ghosh D, Chinnaiyan AM: Meta-analysis of microarrays: interstudy validation of gene expression profiles reveals pathway dysregulation in prostate cancer. Cancer Res. 2002, 62 (15): 4427-4433.
Aerts S, Thijs G, Coessens B, Staes M, Moreau Y, De Moor B: Toucan: deciphering the cis-regulatory logic of coregulated genes. Nucleic Acids Res. 2003, 31 (6): 1753-1764. 10.1093/nar/gkg268.
Wasserman WW, Sandelin A: Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet. 2004, 5 (4): 276-287. 10.1038/nrg1315.
SOURCE search. [http://source.stanford.edu]
NetAffx™ Analysis Center. [http://www.affymetrix.com/analysis/index.affx]
UniGene home. [http://www.ncbi.nlm.nih.gov/unigene]
Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA: DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4 (5): P3-10.1186/gb-2003-4-5-p3.
Chang JT, Nevins JR: GATHER: a systems approach to interpreting genomic signatures. Bioinformatics. 2006, 22 (23): 2926-2933. 10.1093/bioinformatics/btl483.
Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M: From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006, 34 (Database issue): D354-357. 10.1093/nar/gkj102.
BioCarta. [http://www.biocarta.com/genes/index.asp]
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al: Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000, 25 (1): 25-29. 10.1038/75556.
INCLUSive. [http://homes.esat.kuleuven.be/~sistawww/bioi/thijs/download.html]
Wingender E, Chen X, Fricke E, Geffers R, Hehl R, Liebich I, Krull M, Matys V, Michael H, Ohnhauser R, et al: The TRANSFAC system on gene expression regulation. Nucleic Acids Res. 2001, 29 (1): 281-283. 10.1093/nar/29.1.281.
Zheng J, Wu J, Sun Z: An approach to identify over-represented cis-elements in related sequences. Nucleic Acids Research. 2003, 31 (7): 1995-2005. 10.1093/nar/gkg287.
Over-represented Transcription Factor Binding Site Prediction Tool. [http://www.bioinfo.tsinghua.edu.cn/~zhengjsh/OTFBS/]
Quandt K, Frech K, Karas H, Wingender E, Werner T: MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995, 23 (23): 4878-4884. 10.1093/nar/23.23.4878.
Cole SW, Yan W, Galic Z, Arevalo J, Zack JA: Expression-based monitoring of transcription factor activity: the TELiS database. Bioinformatics. 2005, 21 (6): 803-810. 10.1093/bioinformatics/bti038.
Transcription Element Listening System. [http://www.telis.ucla.edu/]
Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B: JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004, 32 (Database issue): D91-94. 10.1093/nar/gkh012.
Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, et al: The UCSC Genome Browser Database. Nucleic Acids Res. 2003, 31 (1): 51-54. 10.1093/nar/gkg129.
Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al: TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003, 31 (1): 374-378. 10.1093/nar/gkg108.
Allegra A, Marino A, Coffaro F, Lama A, Rizza G, Scaglione P, Sammartano F, Santoro A, Volpes A: Is there a uniform basal endometrial gene expression profile during the implantation window in women who became pregnant in a subsequent ICSI cycle?. Hum Reprod. 2009, 24 (10): 2549-2557. 10.1093/humrep/dep222.
Kao LC, Germeyer A, Tulac S, Lobo S, Yang JP, Taylor RN, Osteen K, Lessey BA, Giudice LC: Expression profiling of endometrium from women with endometriosis reveals candidate genes for disease-based implantation failure and infertility. Endocrinology. 2003, 144 (7): 2870-2881. 10.1210/en.2003-0043.
Burney RO, Talbi S, Hamilton AE, Vo KC, Nyegaard M, Nezhat CR, Lessey BA, Giudice LC: Gene expression analysis of endometrium reveals progesterone resistance and candidate susceptibility genes in women with endometriosis. Endocrinology. 2007, 148 (8): 3814-3826. 10.1210/en.2006-1692.
Tapia A, Gangi LM, Zegers-Hochschild F, Balmaceda J, Pommer R, Trejo L, Pacheco IM, Salvatierra AM, Henriquez S, Quezada M, et al: Differences in the endometrial transcript profile during the receptive period between women who were refractory to implantation and those who achieved pregnancy. Hum Reprod. 2008, 23 (2): 340-351. 10.1093/humrep/dem319.
Altmäe S, Martinez-Conejero JA, Salumets A, Simon C, Horcajadas JA, Stavreus-Evers A: Endometrial gene expression analysis at the time of embryo implantation in women with unexplained infertility. Mol Hum Reprod. 2010, 16 (3): 178-187.
Koler M, Achache H, Tsafrir A, Smith Y, Revel A, Reich R: Disrupted gene pattern in patients with repeated in vitro fertilization (IVF) failure. Hum Reprod. 2009, 24 (10): 2541-2548. 10.1093/humrep/dep193.
Catalano RD, Critchley HO, Heikinheimo O, Baird DT, Hapangama D, Sherwin JR, Charnock-Jones DS, Smith SK, Sharkey AM: Mifepristone induced progesterone withdrawal reveals novel regulatory pathways in human endometrium. Mol Hum Reprod. 2007, 13 (9): 641-654. 10.1093/molehr/gam021.
Horcajadas JA, Sharkey AM, Catalano RD, Sherwin JR, Dominguez F, Burgos LA, Castro A, Peraza MR, Pellicer A, Simon C: Effect of an intrauterine device on the gene expression profile of the endometrium. J Clin Endocrinol Metab. 2006, 91 (8): 3199-3207. 10.1210/jc.2006-0430.
Horcajadas JA, Riesewijk A, Dominguez F, Cervero A, Pellicer A, Simon C: Determinants of endometrial receptivity. Ann N Y Acad Sci. 2004, 1034: 166-175. 10.1196/annals.1335.019.
Wang H, Dey SK: Roadmap to embryo implantation: clues from mouse models. Nat Rev Genet. 2006, 7 (3): 185-199. 10.1038/nrg1808.
Horcajadas JA, Pellicer A, Simon C: Wide genomic analysis of human endometrial receptivity: new times, new opportunities. Hum Reprod Update. 2007, 13 (1): 77-86. 10.1093/humupd/dml046.
Lessey BA, Killam AP, Metzger DA, Haney AF, Greene GL, McCarty KS: Immunohistochemical analysis of human uterine estrogen and progesterone receptors throughout the menstrual cycle. J Clin Endocrinol Metab. 1988, 67 (2): 334-340. 10.1210/jcem-67-2-334.
Franchi A, Zaret J, Zhang X, Bocca S, Oehninger S: Expression of immunomodulatory genes, their protein products and specific ligands/receptors during the window of implantation in the human endometrium. Mol Hum Reprod. 2008, 14 (7): 413-421. 10.1093/molehr/gan029.
Janeway CA, Medzhitov R: Innate immune recognition. Annu Rev Immunol. 2002, 20: 197-216. 10.1146/annurev.immunol.20.083001.084359.
Lobo SC, Huang ST, Germeyer A, Dosiou C, Vo KC, Tulac S, Nayak NR, Giudice LC: The immune environment in human endometrium during the window of implantation. Am J Reprod Immunol. 2004, 52 (4): 244-251. 10.1111/j.1600-0897.2004.00217.x.
Hasty LA, Lambris JD, Lessey BA, Pruksananonda K, Lyttle CR: Hormonal regulation of complement components and receptors throughout the menstrual cycle. Am J Obstet Gynecol. 1994, 170 (1 Pt 1): 168-175.
Jensen TS, Bjorge L, Wollen AL, Ulstein M: Identification of the complement regulatory proteins CD46, CD55, and CD59 in human fallopian tube, endometrium, and cervical mucosa and secretion. Am J Reprod Immunol. 1995, 34 (1): 1-9.
Oglesby TJ, Longwith JE, Huettner PC: Human complement regulator expression by the normal female reproductive tract. Anat Rec. 1996, 246 (1): 78-86. 10.1002/(SICI)1097-0185(199609)246:1<78::AID-AR9>3.0.CO;2-B.
Young SL, Lessey BA, Fritz MA, Meyer WR, Murray MJ, Speckman PL, Nowicki BJ: In vivo and in vitro evidence suggest that HB-EGF regulates endometrial expression of human decay-accelerating factor. J Clin Endocrinol Metab. 2002, 87 (3): 1368-1375. 10.1210/jc.87.3.1368.
Nogawa Fonzar-Marana RR, Ferriani RA, Soares SG, Cavalcante-Neto FF, Teixeira JE, Barbosa JE: Expression of complement system regulatory molecules in the endometrium of normal ovulatory and hyperstimulated women correlate with menstrual cycle phase. Fertil Steril. 2006, 86 (3): 758-761. 10.1016/j.fertnstert.2006.02.095.
Isaacson KB, Coutifaris C, Garcia CR, Lyttle CR: Production and secretion of complement component 3 by endometriotic tissue. J Clin Endocrinol Metab. 1989, 69 (5): 1003-1009. 10.1210/jcem-69-5-1003.
Lee J, Oh J, Choi E, Park I, Han C, Kim do H, Choi BC, Kim JW, Cho C: Differentially expressed genes implicated in unexplained recurrent spontaneous abortion. Int J Biochem Cell Biol. 2007, 39 (12): 2265-2277. 10.1016/j.biocel.2007.06.012.
Liu LP, Chan ST, Ho PC, Yeung WS: Partial purification of embryotrophic factors from human oviductal cells. Hum Reprod. 1998, 13 (6): 1613-1619. 10.1093/humrep/13.6.1613.
Xu JS, Cheung TM, Chan ST, Ho PC, Yeung WS: Temporal effect of human oviductal cell and its derived embryotrophic factors on mouse embryo development. Biol Reprod. 2001, 65 (5): 1481-1488. 10.1095/biolreprod65.5.1481.
Lee YL, Lee KF, Xu JS, He QY, Chiu JF, Lee WM, Luk JM, Yeung WS: The embryotrophic activity of oviductal cell-derived complement C3b and iC3b, a novel function of complement protein in reproduction. J Biol Chem. 2004, 279 (13): 12763-12768. 10.1074/jbc.M311160200.
Tse PK, Lee YL, Chow WN, Luk JM, Lee KF, Yeung WS: Preimplantation embryos cooperate with oviductal cells to produce embryotrophic inactivated complement-3b. Endocrinology. 2008, 149 (3): 1268-1276. 10.1210/en.2007-1277.
Giudice LC: Application of functional genomics to primate endometrium: insights into biological processes. Reprod Biol Endocrinol. 2006, 4 (Suppl 1): S4-10.1186/1477-7827-4-S1-S4.
Jackman M, Lindon C, Nigg EA, Pines J: Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat Cell Biol. 2003, 5 (2): 143-148. 10.1038/ncb918.
Jin P, Hardy S, Morgan DO: Nuclear localization of cyclin B1 controls mitotic entry after DNA damage. J Cell Biol. 1998, 141 (4): 875-885. 10.1083/jcb.141.4.875.
Shiozawa T, Li SF, Nakayama K, Nikaido T, Fujii S: Relationship between the expression of cyclins/cyclin-dependent kinases and sex-steroid receptors/Ki67 in normal human endometrial glands and stroma during the menstrual cycle. Mol Hum Reprod. 1996, 2 (10): 745-752. 10.1093/molehr/2.10.745.
Tang L, Zhang Y, Pan H, Luo Q, Zhu XM, Dong MY, Leung PC, Sheng JZ, Huang HF: Involvement of cyclin B1 in progesterone-mediated cell growth inhibition, G2/M cell cycle arrest, and apoptosis in human endometrial cell. Reprod Biol Endocrinol. 2009, 7: 144-10.1186/1477-7827-7-144.
Giguere V: Retinoic acid receptors and cellular retinoid binding proteins: complex interplay in retinoid signaling. Endocr Rev. 1994, 15 (1): 61-79.
Prentice A, Matthews CJ, Thomas EJ, Redfern CP: The expression of retinoic acid receptors in cultured human endometrial stromal cells and effects of retinoic acid. Hum Reprod. 1992, 7 (5): 692-700.
Kumarendran MK, Loughney AD, Prentice A, Thomas EJ, Redfern CP: Nuclear retinoid receptor expression in normal human endometrium throughout the menstrual cycle. Mol Hum Reprod. 1996, 2 (2): 123-129. 10.1093/molehr/2.2.123.
Morriss-Kay GM, Ward SJ: Retinoids and mammalian development. Int Rev Cytol. 1999, 188: 73-131. full_text.
Ito K, Suzuki T, Moriya T, Utsunomiya H, Sugawara A, Konno R, Sato S, Sasano H: Retinoid receptors in the human endometrium and its disorders: a possible modulator of 17 beta-hydroxysteroid dehydrogenase. J Clin Endocrinol Metab. 2001, 86 (6): 2721-2727. 10.1210/jc.86.6.2721.
Zheng WL, Ong DE: Spatial and temporal patterns of expression of cellular retinol-binding protein and cellular retinoic acid-binding proteins in rat uterus during early pregnancy. Biol Reprod. 1998, 58 (4): 963-970. 10.1095/biolreprod58.4.963.
Jeong JW, Lee KY, Kwak I, White LD, Hilsenbeck SG, Lydon JP, DeMayo FJ: Identification of murine uterine genes regulated in a ligand-dependent manner by the progesterone receptor. Endocrinology. 2005, 146 (8): 3490-3505. 10.1210/en.2005-0016.
Fritzsche B, Vermot J, Neumann U, Schmidt A, Schweigert FJ, Dolle P, Ruhl R: Regulation of expression of the retinoic acid metabolizing enzyme CYP26A1 in uteri of ovariectomized mice after treatment with ovarian steroid hormones. Mol Reprod Dev. 2007, 74 (2): 258-264. 10.1002/mrd.20526.
Han BC, Xia HF, Sun J, Yang Y, Peng JP: Retinoic acid-metabolizing enzyme cytochrome P450 26a1 (cyp26a1) is essential for implantation: functional study of its role in early pregnancy. J Cell Physiol. 2010, 223 (2): 471-479.
Deng L, Shipley GL, Loose-Mitchell DS, Stancel GM, Broaddus R, Pickar JH, Davies PJ: Coordinate regulation of the production and signaling of retinoic acid by estrogen in the human endometrium. J Clin Endocrinol Metab. 2003, 88 (5): 2157-2163. 10.1210/jc.2002-021844.
Guzeloglu-Kayisli O, Kayisli UA, Taylor HS: The role of growth factors and cytokines during implantation: endocrine and paracrine interactions. Semin Reprod Med. 2009, 27 (1): 62-79. 10.1055/s-0028-1108011.
Cameron IT, Plumpton C, Champeney R, van Papendorp C, Ashby MJ, Davenport AP: Identification of endothelin-1, endothelin-2 and endothelin-3 in human endometrium. J Reprod Fertil. 1993, 98 (1): 251-255. 10.1530/jrf.0.0980251.
Cameron IT, Davenport AP, van Papendorp C, Barker PJ, Huskisson NS, Gilmour RS, Brown MJ, Smith SK: Endothelin-like immunoreactivity in human endometrium. J Reprod Fertil. 1992, 95 (2): 623-628. 10.1530/jrf.0.0950623.
Shinmi O, Kimura S, Sawamura T, Sugita Y, Yoshizawa T, Uchiyama Y, Yanagisawa M, Goto K, Masaki T, Kanazawa I: Endothelin-3 is a novel neuropeptide: isolation and sequence determination of endothelin-1 and endothelin-3 in porcine brain. Biochem Biophys Res Commun. 1989, 164 (1): 587-593. 10.1016/0006-291X(89)91760-9.
Matsumoto H, Suzuki N, Onda H, Fujino M: Abundance of endothelin-3 in rat intestine, pituitary gland and brain. Biochem Biophys Res Commun. 1989, 164 (1): 74-80. 10.1016/0006-291X(89)91684-7.
Baynash AG, Hosoda K, Giaid A, Richardson JA, Emoto N, Hammer RE, Yanagisawa M: Interaction of endothelin-3 with endothelin-B receptor is essential for development of epidermal melanocytes and enteric neurons. Cell. 1994, 79 (7): 1277-1285. 10.1016/0092-8674(94)90018-3.
Montani D, Souza R, Binkert C, Fischli W, Simonneau G, Clozel M, Humbert M: Endothelin-1/endothelin-3 ratio: a potential prognostic factor of pulmonary arterial hypertension. Chest. 2007, 131 (1): 101-108. 10.1378/chest.06-0682.
Jeoung M, Lee S, Hawng HK, Cheon YP, Jeong YK, Gye MC, Iglarz M, Ko C, Bridges PJ: Identification of a novel role for endothelins within the oviduct. Endocrinology. 2010, 151 (6): 2858-2867. 10.1210/en.2009-1155.
Ferriani RA, Charnock-Jones DS, Prentice A, Thomas EJ, Smith SK: Immunohistochemical localization of acidic and basic fibroblast growth factors in normal human endometrium and endometriosis and the detection of their mRNA by polymerase chain reaction. Hum Reprod. 1993, 8 (1): 11-16.
Anania CA, Stewart EA, Quade BJ, Hill JA, Nowak RA: Expression of the fibroblast growth factor receptor in women with leiomyomas and abnormal uterine bleeding. Mol Hum Reprod. 1997, 3 (8): 685-691. 10.1093/molehr/3.8.685.
Sangha RK, Li XF, Shams M, Ahmed A: Fibroblast growth factor receptor-1 is a critical component for endometrial remodeling: localization and expression of basic fibroblast growth factor and FGF-R1 in human endometrium during the menstrual cycle and decreased FGF-R1 expression in menorrhagia. Lab Invest. 1997, 77 (4): 389-402.
Moller B, Rasmussen C, Lindblom B, Olovsson M: Expression of the angiogenic growth factors VEGF, FGF-2, EGF and their receptors in normal human endometrium during the menstrual cycle. Mol Hum Reprod. 2001, 7 (1): 65-72. 10.1093/molehr/7.1.65.
Irwin JC, Utian WH, Eckert RL: Sex steroids and growth factors differentially regulate the growth and differentiation of cultured human endometrial stromal cells. Endocrinology. 1991, 129 (5): 2385-2392. 10.1210/endo-129-5-2385.
Presta M: Sex hormones modulate the synthesis of basic fibroblast growth factor in human endometrial adenocarcinoma cells: implications for the neovascularization of normal and neoplastic endometrium. J Cell Physiol. 1988, 137 (3): 593-597. 10.1002/jcp.1041370329.
Satterfield MC, Song G, Hayashi K, Bazer FW, Spencer TE: Progesterone regulation of the endometrial WNT system in the ovine uterus. Reprod Fertil Dev. 2008, 20 (8): 935-946. 10.1071/RD08069.
Pavlova A, Boutin E, Cunha G, Sassoon D: Msx1 (Hox-7.1) in the adult mouse uterus: cellular interactions underlying regulation of expression. Development. 1994, 120 (2): 335-345.
Daikoku T, Song H, Guo Y, Riesewijk A, Mosselman S, Das SK, Dey SK: Uterine Msx-1 and Wnt4 signaling becomes aberrant in mice with the loss of leukemia inhibitory factor or Hoxa-10: evidence for a novel cytokine-homeobox-Wnt signaling in implantation. Mol Endocrinol. 2004, 18 (5): 1238-1250. 10.1210/me.2003-0403.
Mansouri-Attia N, Sandra O, Aubert J, Degrelle S, Everts RE, Giraud-Delville C, Heyman Y, Galio L, Hue I, Yang X, et al: Endometrium as an early sensor of in vitro embryo manipulation technologies. Proc Natl Acad Sci USA. 2009, 106 (14): 5687-5692. 10.1073/pnas.0812722106.
Norton JD: ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J Cell Sci. 2000, 113 (Pt 22): 3897-3905.
Yokota Y: Id and development. Oncogene. 2001, 20 (58): 8290-8298. 10.1038/sj.onc.1205090.
Lasorella A, Uo T, Iavarone A: Id proteins at the cross-road of development and cancer. Oncogene. 2001, 20 (58): 8326-8333. 10.1038/sj.onc.1205093.
Benezra R, Rafii S, Lyden D: The Id proteins and angiogenesis. Oncogene. 2001, 20 (58): 8334-8341. 10.1038/sj.onc.1205160.
Maldonado V, Castilla JA, Martinez L, Herruzo A, Concha A, Fontes J, Mendoza N, Garcia-Pena ML, Mendoza JL, Magan R, et al: Expression of transcription factors in endometrium during natural cycles. J Assist Reprod Genet. 2003, 20 (11): 474-481. 10.1023/B:JARG.0000006710.64788.99.
Kilpatrick LM, Kola I, Salamonsen LA: Transcription factors Ets1, Ets2, and Elf1 exhibit differential localization in human endometrium across the menstrual cycle and alternate isoforms in cultured endometrial cells. Biol Reprod. 1999, 61 (1): 120-126. 10.1095/biolreprod61.1.120.
Christian M, Zhang X, Schneider-Merck T, Unterman TG, Gellersen B, White JO, Brosens JJ: Cyclic AMP-induced forkhead transcription factor, FKHR, cooperates with CCAAT/enhancer-binding protein beta in differentiating human endometrial stromal cells. J Biol Chem. 2002, 277 (23): 20825-20832. 10.1074/jbc.M201018200.
Mak IY, Brosens JJ, Christian M, Hills FA, Chamley L, Regan L, White JO: Regulated expression of signal transducer and activator of transcription, Stat5, and its enhancement of PRL expression in human endometrial stromal cells in vitro. J Clin Endocrinol Metab. 2002, 87 (6): 2581-2588. 10.1210/jc.87.6.2581.
Martin R, Taylor MB, Krikun G, Lockwood C, Akbas GE, Taylor HS: Differential cell-specific modulation of HOXA10 by estrogen and specificity protein 1 response elements. J Clin Endocrinol Metab. 2007, 92 (5): 1920-1926. 10.1210/jc.2006-1694.
Lynch VJ, Brayer K, Gellersen B, Wagner GP: HoxA-11 and FOXO1A cooperate to regulate decidual prolactin expression: towards inferring the core transcriptional regulators of decidual genes. PLoS One. 2009, 4 (9): e6845-10.1371/journal.pone.0006845.
Kohan K, Carvajal R, Gabler F, Vantman D, Romero C, Vega M: Role of the transcriptional factors FOXO1 and PPARG on gene expression of SLC2A4 in endometrial tissue from women with polycystic ovary syndrome. Reproduction. 2010, 140 (1): 123-131. 10.1530/REP-10-0056.
Fahnenstich J, Nandy A, Milde-Langosch K, Schneider-Merck T, Walther N, Gellersen B: Promyelocytic leukaemia zinc finger protein (PLZF) is a glucocorticoid- and progesterone-induced transcription factor in human endometrial stromal cells and myometrial smooth muscle cells. Mol Hum Reprod. 2003, 9 (10): 611-623. 10.1093/molehr/gag080.
Pohnke Y, Schneider-Merck T, Fahnenstich J, Kempf R, Christian M, Milde-Langosch K, Brosens JJ, Gellersen B: Wild-type p53 protein is up-regulated upon cyclic adenosine monophosphate-induced differentiation of human endometrial stromal cells. J Clin Endocrinol Metab. 2004, 89 (10): 5233-5244. 10.1210/jc.2004-0012.
Du H, Sarno J, Taylor HS: HOXA10 inhibits Kruppel-like factor 9 expression in the human endometrial epithelium. Biol Reprod. 2010, 83 (2): 205-211. 10.1095/biolreprod.110.083980.
Kim YA, Kim JY, Kim MR, Hwang KJ, Chang DY, Jeon MK: Tumor necrosis factor-alpha-induced cyclooxygenase-2 overexpression in eutopic endometrium of women with endometriosis by stromal cell culture through nuclear factor-kappaB activation. J Reprod Med. 2009, 54 (10): 625-630.
Pabona JM, Zeng Z, Simmen FA, Simmen RC: Functional differentiation of uterine stromal cells involves cross-regulation between bone morphogenetic protein 2 and Kruppel-like factor (KLF) family members KLF9 and KLF13. Endocrinology. 2010, 151 (7): 3396-3406. 10.1210/en.2009-1370.
Strakova Z, Reed J, Ihnatovych I: Human transcriptional coactivator with PDZ-binding motif (TAZ) is downregulated during decidualization. Biol Reprod. 2010, 82 (6): 1112-1118. 10.1095/biolreprod.109.081844.
Cook IH, Evans J, Maldonado-Perez D, Critchley HO, Sales KJ, Jabbour HN: Prokineticin-1 (PROK1) modulates interleukin (IL)-11 expression via prokineticin receptor 1 (PROKR1) and the calcineurin/NFAT signalling pathway. Mol Hum Reprod. 2010, 16 (3): 158-169. 10.1093/molehr/gap084.
Wang Y, Hanifi-Moghaddam P, Hanekamp EE, Kloosterboer HJ, Franken P, Veldscholte J, van Doorn HC, Ewing PC, Kim JJ, Grootegoed JA, et al: Progesterone inhibition of Wnt/beta-catenin signaling in normal endometrium and endometrial cancer. Clin Cancer Res. 2009, 15 (18): 5784-5793. 10.1158/1078-0432.CCR-09-0814.
Johnson DG, Schneider-Broussard R: Role of E2F in cell cycle control and cancer. Front Biosci. 1998, 3: d447-448.
La Thangue NB: The yin and yang of E2F-1: balancing life and death. Nat Cell Biol. 2003, 5 (7): 587-589. 10.1038/ncb0703-587.
Pierce AM, Schneider-Broussard R, Gimenez-Conti IB, Russell JL, Conti CJ, Johnson DG: E2F1 has both oncogenic and tumor-suppressive properties in a transgenic model. Mol Cell Biol. 1999, 19 (9): 6408-6414.
Murre C, McCaw PS, Baltimore D: A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989, 56 (5): 777-783. 10.1016/0092-8674(89)90682-X.
Benezra R, Davis RL, Lassar A, Tapscott S, Thayer M, Lockshon D, Weintraub H: Id: a negative regulator of helix-loop-helix DNA binding proteins. Control of terminal myogenic differentiation. Ann N Y Acad Sci. 1990, 599: 1-11. 10.1111/j.1749-6632.1990.tb42359.x.
Yokoyama C, Wang X, Briggs MR, Admon A, Wu J, Hua X, Goldstein JL, Brown MS: SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993, 75 (1): 187-197.
Brown MS, Goldstein JL: The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997, 89 (3): 331-340. 10.1016/S0092-8674(00)80213-5.
Brown MS, Goldstein JL: A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci USA. 1999, 96 (20): 11041-11048. 10.1073/pnas.96.20.11041.
Shea-Eaton WK, Trinidad MJ, Lopez D, Nackley A, McLean MP: Sterol regulatory element binding protein-1a regulation of the steroidogenic acute regulatory protein gene. Endocrinology. 2001, 142 (4): 1525-1533. 10.1210/en.142.4.1525.
Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H: The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990, 61 (1): 49-59. 10.1016/0092-8674(90)90214-Y.
Kreider BL, Benezra R, Rovera G, Kadesch T: Inhibition of myeloid differentiation by the helix-loop-helix protein Id. Science. 1992, 255 (5052): 1700-1702. 10.1126/science.1372755.
Moldes M, Boizard M, Liepvre XL, Feve B, Dugail I, Pairault J: Functional antagonism between inhibitor of DNA binding (Id) and adipocyte determination and differentiation factor 1/sterol regulatory element-binding protein-1c (ADD1/SREBP-1c) trans-factors for the regulation of fatty acid synthase promoter in adipocytes. Biochem J. 1999, 344 (Pt 3): 873-880. 10.1042/0264-6021:3440873.
Fontemaggi G, Dell'Orso S, Trisciuoglio D, Shay T, Melucci E, Fazi F, Terrenato I, Mottolese M, Muti P, Domany E, et al: The execution of the transcriptional axis mutant p53, E2F1 and ID4 promotes tumor neo-angiogenesis. Nat Struct Mol Biol. 2009, 16 (10): 1086-1093. 10.1038/nsmb.1669.
Giudice LC: Genes associated with embryonic attachment and implantation and the role of progesterone. J Reprod Med. 1999, 44 (2 Suppl): 165-171.
Tabibzadeh S: Molecular control of the implantation window. Hum Reprod Update. 1998, 4 (5): 465-471. 10.1093/humupd/4.5.465.
O'Malley BW, Tsai MJ: Molecular pathways of steroid receptor action. Biol Reprod. 1992, 46 (2): 163-167.
Wagner BL, Norris JD, Knotts TA, Weigel NL, McDonnell DP: The nuclear corepressors NCoR and SMRT are key regulators of both ligand- and 8-bromo-cyclic AMP-dependent transcriptional activity of the human progesterone receptor. Mol Cell Biol. 1998, 18 (3): 1369-1378.
Archer TK, Deroo BJ, Fryer CJ: Chromatin modulation of glucocorticoid and progesterone receptor activity. Trends Endocrinol Metab. 1997, 8 (10): 384-390. 10.1016/S1043-2760(97)00159-8.
McKenna NJ, O'Malley BW: Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002, 108 (4): 465-474. 10.1016/S0092-8674(02)00641-4.
Fernandes MS, Pierron V, Michalovich D, Astle S, Thornton S, Peltoketo H, Lam EW, Gellersen B, Huhtaniemi I, Allen J, et al: Regulated expression of putative membrane progestin receptor homologues in human endometrium and gestational tissues. J Endocrinol. 2005, 187 (1): 89-101. 10.1677/joe.1.06242.
Zhang L, Kanda Y, Roberts DJ, Ecker JL, Losel R, Wehling M, Peluso JJ, Pru JK: Expression of progesterone receptor membrane component 1 and its partner serpine 1 mRNA binding protein in uterine and placental tissues of the mouse and human. Mol Cell Endocrinol. 2008, 287 (1-2): 81-89. 10.1016/j.mce.2008.02.012.
Wade HE, Kobayashi S, Eaton ML, Jansen MS, Lobenhofer EK, Lupien M, Geistlinger TR, Zhu W, Nevins JR, Brown M, et al: Multimodal regulation of E2F1 gene expression by progestins. Mol Cell Biol. 2010, 30 (8): 1866-1877. 10.1128/MCB.01060-09.
Acknowledgements
We thank Reinaldo González-Ramos and M. Cecilia Johnson (IDIMI, Universidad de Chile, Santiago, Chile) for their helpful comments and critical review of the article. We thank also Lois Salamonsen (Prince Henry's Institute, Melbourne, Australia) for her comments and suggestions. This study was supported by FONDECYT 3090075 and FONDAP-15010006. A.T. is supported by PBCT-PSD51(IDIMI).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
AT conceived of the study, participated in its design, carried out the bioinformatic analyses and helped to draft the manuscript. CV participated in the creation and intersection of database. JCM participated in its design and data analysis. HBC participated in the coordination and helped to draft the manuscript. LD contributed to data analysis and helped to draft the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Tapia, A., Vilos, C., Marín, J.C. et al. Bioinformatic detection of E47, E2F1 and SREBP1 transcription factors as potential regulators of genes associated to acquisition of endometrial receptivity. Reprod Biol Endocrinol 9, 14 (2011). https://doi.org/10.1186/1477-7827-9-14
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1477-7827-9-14