Abstract
Background
Exposure to dioxins results in a broad range of pathophysiological disorders in human fetuses. In order to evaluate the effects of dioxins on the feto-placental tissues, we analyzed the gene expression in 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) treated primary cultures of human amniotic epithelial cells.
Methods
Human amniotic epithelial cells were dispersed by trypsin from amniotic membranes and cultured in DME/Ham's F12 medium supplemented with 10% FBS. Two weeks after plating, cells were treated with 50 nM TCDD or DMSO (control), further incubated for 48 hrs, and the gene expression was analyzed by DNA microarray technology and quantitative real-time PCR.
Results
Thirty eight TCDD-inducible genes, including cytochromeP4501A1 and cytochromeP4501B1, were identified. One of the remarkable profiles of the gene expression was the prominent up-regulation of interferon-inducible genes. The genes involved in the interferon gene expression and interferon signaling pathways were also up-regulated. Furthermore, the expression of genes related to collagen synthesis or degradation was enhanced by TCDD.
Conclusion
Using DNA microarray and quantitative real-time PCR analyses, we identified TCDD-inducible genes, including interferon-inducible genes and genes related to collagen synthesis or degradation, in human amniotic epithelial cells.
Similar content being viewed by others
Background
Exposure to dioxins causes a diverse spectrum of toxicities in humans and laboratory animals [1–4]. The fetus is one of the most sensitive targets of dioxins and a broad range of pathophysiological abnormalities, such as, disorders of brain development, thyroxin resistance, hepatic damage, hematopoietic disorders and lung dysfunction, are observed in humans after perinatal exposure to dioxins [1]. Dioxins are transferred to fetuses and infants through placentas and milk from mothers [5]. Dioxins were detected in all of the samples analyzed in a study performed using human umbilical cord or cord serum in Japan [6]. Higher dioxin levels were reported in the placenta compared to that in breast milk, in a study performed in Taiwan [7]. Not only the morphological and functional disorders brought about by the altered gene products but the comprehensive analyses of the change in gene expression are required to evaluate the effects of fetal exposure to endocrine disruptors [8]. However those studies of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) on human feto-placentas are limited [6–13].
The amniotic membrane is of fetal origin but contains no blood vessels [14]. The amniotic epithelial cells (AEC) on the basement membrane which is the thickest among basement membranes in human lay in the most interior of the fetal membranes. From the histological character mentioned above, it is relatively easier to obtain homogenous cells for experiments without contamination of hematocytes or cells of blood vessel origin [15]. Moreover, since fetal membranes are the tissues disposed of after parturition, the ethical issues associated with use for experiments are limited. Therefore, we analyzed gene expression induced by TCDD, the most toxic congener among the dioxins, in primary cultures of human AEC using DNA microarray technology, a powerful tool in revealing global gene expression, and quantitative real-time PCR.
Methods
Reagents
2,3,7,8-TCDD was obtained from Cambridge Isotope Laboratory (Andover, MA). Human oligonucleotide glass array (Human Genome U133A Array) was from Affymetrix (Santa Clara, CA).
AEC culture and TCDD treatment
With the permission of The Internal Review Board for Gunma University Hospital and the written informed consent of the patients, we obtained fetal membrane samples from patients with single pregnancy without systemic disease, signs of premature delivery or fetal complications, during elective cesarean sections at term. Three fetal membranes from three women were used for independent primary cultures. AEC were prepared based on the method described by Okita et al [15] with slight modifications, after an amniotic membrane was mechanically peeled off from the chorion. Briefly, the membrane was incubated in 170 ml of Krebs-Ringer solution containing 0.15 % trypsin, 1.26 g/l sodium bicarbonate, 25 mM HEPES, 50 μg/ml streptomycin and 0.25 μg/ml amphotericin B at 37C using a Spinner Flask. The liberated cells were decanted at 30-min intervals and the incubation was performed seven times with freshly made trypsin solution. Each fraction of dispersed cells were centrifuged, resuspended in DME/Ham's F12 medium supplemented with 15 mM HEPES, 10% FBS, 50 μg/ml streptomycin and 0.25 μg/ml amphotericin B. The first fraction was discarded. The cell viability of the remaining fractions was determined by trypan blue exclusion and the fractions with viabilities not less than 80% were pooled. Cells were plated at the density of 2 × 106 cells per 25 cm2 polystyrene flask in 10 ml of medium and cultured in a humidified atmosphere containing 5% CO2 – 95% air at 37C. The medium was changed every 2 to 3 days. The experiments were performed two weeks after plating when cells were sub-confluent to confluent. The cells were treated with 50 nM TCDD or DMSO (control), and further incubated for 48 hrs. Total RNA from these cells was isolated and used for DNA microarray and quantitative Real-Time PCR analyses.
DNA microarray
The microarray method was carried out according to the manufacture's instruction. Briefly, total RNA was extracted from the TCDD-treated cells and control AEC using an RNA extraction solution (Trizol). After Oligo-dT latex beads treatment, double stranded cDNA libraries were constructed from the mRNA of TCDD-treated cells and the control AEC using an oligo-dT primer with a T7-promoter sequence at the 5'-end. Biotin-labeled complementary RNA was in vitro transcribed by T7 promoter using the cDNA libraries as template. The biotin-labeled RNA was fragmented. Each sample was hybridized to a separate oligonucleotide array (Affymetrix Human Genome U133A) for 16 hours at 45C, and then washed and stained with streptavidin phycoerythrin conjugate using GeneChip Fluidics. The arrays were scanned by a Gene-Array scanner using Affymetrix GeneChip Microarray Suite (MAS) 5.0 software for scanning and basic analysis.
The Human Genome U133A array contains 22,277 probe sets including 61 control probe sets, and analyzes the expression level of 18,720 full length transcripts with 13,900 characterized human genes. The arrays incorporate a perfect match (PM)/mismatch (MM) probe pair strategy. One probe from the pair perfectly matches its target sequence while the other contains a mismatch located at the center of the 25-mer sequence. This probe pairing design helps identify and subtract non-specific hybridization and background signal. For each probe pair, the MM signal was subtracted from the PM signal. The average of these differences was reflective of the level of expression of the gene. The global method of scaling/normalization was performed by MAS 5.0 software. A program (PathwayFinder) was used for pathway analysis. Three independent experiments were performed.
Quantitative real-time PCR
Messenger RNA was extracted using an RNA extraction solution (Trizol) and oligo-dT latex beads as described previously [15, 16]. Five micrograms of mRNA preparations were reverse-transcribed, and then converted to double stranded cDNA molecules. Complementary DNA was quantified by UV absorption measurement, and 1 ng was subjected to the PCR reaction as template. As an internal standard, TATA binding protein (TBP) was used instead of GAPDH, since GAPDH gene expression was affected by the TCDD treatment (data not shown). Quantitative real-time PCR was carried out in an ABI PRISM® 7000 Sequence Detection System (Applied Biosystems; Foster City, CA), according to the manufacture's instructions. PCR reaction involved template cDNA samples, Advantage Taq Plus DNA polymerase (Clontech Laboratories, Inc.; Mountain View, CA), dNTP, and Syber Green. The thermal cycling conditions included an initial incubation of samples at 94°C for 2 min, followed by appropriate cycles of 94°C for 20 sec, 54°C for 30 sec and 72°C for 45 sec. Syber Green fluorescence was used to detect the amplified products. Serial dilutions of the templates were used to create a concentration curve, and relative expression levels were calculated using TBP as normalization control for each sample. Abundance of each gene was referred to as a Ct value in this system [16, 17]. Three independent experiments using AEC from three women were performed. Comparisons between the genes were performed using one-way ANOVA. The significance of the differences between the mean values of cytochromeP4501A1 (CYP1A1) and each gene was tested using paired t-test. P < 0.05 was considered statistically significant.
Results
Among the 22,277 genes spotted on the array, 12,509 genes were expressed in AEC population. Six hundred and ninety six TCDD-sensitive candidate genes were picked up; 326 were TCDD-inducible and 370 were TCDD-suppressive candidate genes (cut-off values of 1.9 as inducible and 0.5 as suppressive genes). Specific primers for 55 genes among 696 TCDD-sensitive candidate genes were synthesized (Table 1). Those genes were selected for two reasons. One reason was that TCDD-sensitive genes in rat placenta had been examined previously [16]. In order to reveal the relationship of sensitive genes between rat placenta and AEC, those genes were selected. Another reason was that even the candidate genes might not represent actual sensitive genes, because false positive signals could not be excluded from the array data, especially when induction or suppression ratios were near 1.9 or 0.5. Therefore we selected genes with higher ratios for real-time PCR.
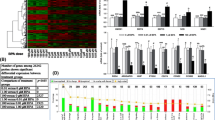
Genes that showed expression ratios (TCDD-treated/control) of more than 1.9 or less than 0.6 were finally identified as TCDD-sensitive. Among 55 genes analyzed, 38 genes were TCDD-inducible (Table 2), 4 were -suppressive (Table 3), and 13 were neither of them, by both DNA microarray technology and quantitative real-time PCR. TCDD-inducible genes were categorized into several groups. Enzyme genes include CYP1A1 and cytochromeP4501B1 (CYP1B1) which are known as the typical TCDD target genes. One of the remarkable findings of the present experiment was that many interferon-related genes were induced in TCDD-treated human AEC (Figure 1). The expression of interferon-inducible genes, that is, interferon induced transmembrane protein 1 (IFITM1), interferon, alpha-inducible protein (G1P2), interferon, alpha-inducible protein 27 (IFI27) and interferon-induced protein with tetratricopeptide repeats 1 (IFIT1) was strongly enhanced. The genes involved in the interferon gene expression and signaling pathway, that is, interferon regulatory factor 7 (IFR7), interferon-stimulated transcription factor 3, gamma 48 kDa (ISGF3G) and signal transducer and activator of transcription 1 (STAT1), were also up-regulated.
Induction of CYP1A1, CYP1B1 and interferon related genes in human AEC by TCDD. Expression levels were determined by DNA microarray and quantitative real-time PCR analyses as described in Methods. Values were expressed as fold induction (TCDD/control). The representative microarray data are shown as yellow columns. The values obtained by PCR analyses are shown as red columns. The data are shown as mean ± SD of three independent experiments. *, P < 0.05 compared with the value of CYP1A1.
The other aspect in TCDD-treated human AEC was that integrin, alpha 10 (ITGA10), integrin, alpha 2 (ITGA2), and matrix metalloproteinase 9 (MMP9), which were genes involved in collagen synthesis and degradation, were also induced by TCDD treatment. However no changes in cellular morphology, number and viability were observed under the experimental conditions.
Discussion
The amniotic membrane composes the innermost layer, nearest the fetus, of the fetal membranes containing the fetus and amniotic fluid [14]. It contains no blood vessels or nerves; the nutrients it requires are supplied by the amniotic fluid. The amniotic epithelium derived from epiblasts of embryo, laid in the most interior of the amniotic membrane, is in contact with amniotic fluid directly. The DNA microarray is a powerful and comprehensive method to identify inducible or suppressive genes by a given hormonal or pharmacological treatment. Using DNA microarray and quantitative real-time PCR technology, we analyzed TCDD-inducible genes in primary cultures of human AEC.
We measured gene expression after 48 hour incubation. Therefore early induced genes are not included in the present study. Biological persistence of TCDD is well known. It is reported that the expression of CYP1A1 mRNA was shown after 6 hours and sustained for up to 72 hours in human colon carcinoma cells treated with TCDD [18]. The maximum effect of TCDD on the expression of FSH receptor mRNA in rat granulosa cell cultures was shown after a 48 hour-incubation in our previous study [19]. The effect of TCDD on the expression of LH receptor mRNA was shown after 24 hours and sustained for up to 72 hours in the same cells [20]. Therefore, we studied TCDD-inducible genes in human AEC after a 48 hour-incubation.
The most striking observation in this experiment was that many interferon-inducible genes were prominently up-regulated in TCDD-treated human AEC; the increase of gene expression of IFITM1, G1P2, IFI27 and IFIT1 was equal to or far more than that of CYP1A1 and CYP1B1 which are well known as TCDD-inducible genes. The genes involved in the interferon gene expression and signaling pathway, that is, IFR7, ISGF3G and STAT1, were also up-regulated. The induction of interferon-related genes by TCDD was first reported by Mizutani et al., in the placentas of TCDD-treated Holtzman rats [16]. The present study confirmed that TCDD induces interferon-related genes in cells derived from human fetuses.
On the other hand, the up-regulation of glucose transporter genes was not observed in AEC culture by DNA microarray analysis (data are not shown), though they were strongly up-regulated in TCDD-treated rat placentas [16]. The placenta is the organ rich in blood vessels and it is reported that exposure to TCDD causes a hypoxic state in the placentas and the glucose kinetics are also altered in those organs [21, 22]. On the other hand, interferon is known to be involved in the regulation of angiogenesis. Mizutani et al speculated that the activation of the interferon signaling pathway impaired the angiogenesis in TCDD-treated rat placentas and brought about a hypoxic state in the placentas which up-regulated glucose transporter gene expression [16]. It was deduced that the expression of glucose transporter genes was not up-regulated in AEC, since both the control and TCDD-treated cells had an adequate supply of oxygen when in the in vitro culture condition. AEC cultures in the present study were performed under air conditions. In future studies, comparing TCDD-inducible genes between AEC cultures under hypoxic conditions and under air conditions would verify the inference and show which condition is closer to in vivo.
Although no changes in cellular morphology, number and viability were observed under the experimental conditions, it was an interesting observation that the expression of genes related to the synthesis and degradation of collagen, that is, MMP9, ITGA2 and ITGA10 were also up-regulated by TCDD in human AEC. Martinez JM et al reported the induction of MMP1, MMM9 and tissue inhibitor of metalloproteinase 3 in TCDD-treated human airway epithelial cells using microarray analysis in 2002 [23]. Thereafter, increased mRNA levels of MMPs were reported in TCDD-treated human cells, such as, MMP1 in keratinocytes [24] and melanoma cells [25], MMP2 in melanoma cells [25], MMP3 and MMP7 in endometrial cells [26], and MMP9 in prostate cancer cells [27] and melanoma cells [25]. Although the mechanisms of induction of MMP genes by TCDD are not fully clarified, Murphy et al reported that TCDD activation of MMP1 mRNA expression was mediated through increased promoter activity [24]. Villano CM et al reported aryl hydrocarbon receptor dependent MMP1, 2, and 9 expression by TCDD and speculated that MMP expression may be a common endpoint for activation of the aryl hydrocarbon receptor pathway [25]. It is speculated that TCDD caused pathological lesions by altering the expression of genes involved in matrix remodeling [24]. On the other hand, increased premature labor is reported in women exposed to dioxins in Chapaevsk [28]. The tensile strength of fetal membranes is provided almost exclusively by the amnion, and the interstitial collagens are believed to be the major source of the tensile strength of this tissue. AEC secrete not only collagens and noncollagenous glycoproteins but also MMPs [14]. Considering these biological characters of the amnion and the present observations on increased expression of genes related to the synthesis and degradation of collagens, our findings permit a better understanding of the pathology of premature labor by dioxins. The study of mechanisms on the induction of these genes by TCDD in human AEC would be the subject of future work.
The data of TCDD-inducible and -suppressive genes in human AEC culture is available at ED-Genes [29].
Conclusion
We identified 38 TCDD-inducible genes in human AEC using DNA microarray and quantitative real-time PCR analyses. Interferon-inducible genes and genes related to collagen synthesis or degradation were up-regulated. These observations indicate that the comprehensive methods used in this study are useful in investigating the effects of dioxins on human feto-placenta.
References
ten Tusscher GW, Koppe JG: Perinatal dioxin exposure and later effects – a review. Chemosphere. 2004, 54: 1329-1336. 10.1016/S0045-6535(03)00254-6.
Mandal PK: Dioxin: a review of its environmental effects and its aryl hydrocarbon receptor biology. J Comp Physiol [B]. 2005, 175: 221-230.
Kogevinas M: Human health effects of dioxins: cancer, reproductive and endocrine system effects. Hum Reprod Update. 2001, 7: 331-339. 10.1093/humupd/7.3.331.
Couture LA, Abbott BD, Birnbaum LS: A critical review of the developmental toxicity and teratogenicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin: recent advances toward understanding the mechanism. Teratology. 1990, 42: 619-627. 10.1002/tera.1420420606.
Przyrembel H, Heinrich-Hirsch B, Vieth B: Exposition to and health effects of residues in human milk. Adv Exp Med Biol. 2000, 478: 307-325.
Todaka E, Mori C: Necessity to establish new risk assessment and risk communication for human fetal exposure to multiple endocrine disruptors in Japan. Congenital Anomalies. 2002, 42: 87-93.
Wang SL, Lin CY, Guo YL, Lin LY, Chou WL, Chang LW: Infant exposure to polychlorinated dibenzo-p-dioxins, dibenzofurans and biphenyls (PCDD/Fs, PCBs) – correlation between prenatal and postnatal exposure. Chemosphere. 2004, 54: 1459-1473. 10.1016/j.chemosphere.2003.08.012.
Mori C, Komiyama M, Adachi T, Sakurai K, Nishimura D, Takashima K, Todaka E: Application of toxicogenomic analysis to risk assessment of delayed long-term effects of multiple chemicals including endocrine disruptors in human fetuses. Environ Health Perspect. 2003, 111: 803-809.
Okey AB, Giannone JV, Smart W, Wong JM, Manchester DK, Parker NB, Feeley MM, Grant DL, Gilman A: Binding of 2,3,7,8-tetrachlorodibenzo-p-dioxin to AH receptor in placentas from normal versus abnormal pregnancy outcomes. Chemosphere. 1997, 34: 1535-1547. 10.1016/S0045-6535(97)00449-9.
Chen J, Thirkill TL, Overstreet JW, Lasley BL, Douglas GC: Effect of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) on chorionic gonadotropin secretion by human trophoblasts. Reprod Toxicol. 2003, 17: 87-93. 10.1016/S0890-6238(02)00077-1.
Augustowska K, Gregoraszczuk EL, Milewicz T, Krzysiek J, Grochowalski A, Chrzaszcz R: Effects of dioxin (2,3,7,8-TCDD) and PCDDs/PCDFs congeners mixture on steroidogenesis in human placenta tissue culture. Endocr Regul. 2003, 37: 11-19.
Augustowska K, Gregoraszczuk EEL, Grochowalski A, Milewicz T, Mika M, Krzysiek J, Chrzaszcz R: Comparison of accumulation and altered steroid secretion by placental tissue treated with TCDD and natural mixture of PCDDs-PCDFs. Reproduction. 2003, 126: 681-687. 10.1530/rep.0.1260681.
Yamamoto J, Ihara K, Nakayama H, Hikino S, Satoh K, Kubo N, Iida T, Fujii Y, Hara T: Characteristic expression of aryl hydrocarbon receptor repressor gene in human tissues: organ-specific distribution and variable induction patterns in mononuclear cells. Life Sci. 2004, 74: 1039-1049. 10.1016/j.lfs.2003.07.022.
Parry S, Strauss JF: Premature rupture of the fetal membranes. N Engl J Med. 1998, 338: 663-670. 10.1056/NEJM199803053381006.
Okita JR, Sagawa N, Casey ML, Snyder JM: A comparison of human amnion tissue and amnion cells in primary culture by morphological and biochemical criteria. In Vitro. 1983, 19: 117-126.
Mizutani T, Yoshino M, Satake T, Nakagawa M, Ishimura R, Tohyama C, Kokame K, Kangawa K, Miyamoto K: Identification of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-inducible and -suppressive genes in the rat placenta: induction of interferon-regulated genes with possible inhibitory roles for angiogenesis in the placenta. Endocr J. 2004, 51: 569-577. 10.1507/endocrj.51.569.
Lancaster JM, Dressman HK, Whitaker RS, Havrilesky L, Gray J, Marks JR, Nevins JR, Berchuck A: Gene expression patterns that characterize advanced stage serous ovarian cancers. J Soc Gynecol Investig. 2004, 11: 51-59. 10.1016/j.jsgi.2003.07.004.
Li W, Harper PA, Tang BK, Okey AB: Regulation of cytochrome P450 enzymes by aryl hydrocarbon receptor in human cells: CYP1A2 expression in the LS180 colon carcinoma cell line after treatment with 2,3,7,8-tetrachlorodibenzo-p-dioxin or 3-methylcholanthrene. Biochem Pharmacol. 1998, 56 (5): 599-612. 10.1016/S0006-2952(98)00208-1.
Hirakawa T, Minegishi T, Abe K, Kishi H, Inoue K, Ibuki Y, Miyamoto K: Effect of 2,3,7,8-tetrachlorodibenzo-p-dioxin on the expression of follicle-stimulating hormone receptors during cell differentiation in cultured granulosa cells. Endocrinology. 2000, 141 (4): 1470-1476. 10.1210/en.141.4.1470.
Hirakawa T, Minegishi T, Abe K, Kishi H, Ibuki Y, Miyamoto K: Effect of 2,3,7,8-tetrachlorodibenzo-p-dioxin on the expression of luteinizing hormone receptors during cell differentiation in cultured granulosa cells. Arch Biochem Biophys. 2000, 375 (2): 371-376. 10.1006/abbi.1999.1678.
Ishimura R, Ohsako S, Kawakami T, Sakaue M, Aoki Y, Tohyama C: Altered protein profile and possible hypoxia in the placenta of 2,3,7,8-tetrachlorodibenzo-p-dioxin-exposed rats. Toxicol Appl Pharmacol. 2002, 185: 197-206. 10.1006/taap.2002.9539.
Ishimura R, Ohsako S, Miyabara Y, Sakaue M, Kawakami T, Aoki Y, Yonemoto J, Tohyama C: Increased glycogen content and glucose transporter 3 mRNA level in the placenta of Holtzman rats after exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Appl Pharmacol. 2002, 178: 161-171. 10.1006/taap.2001.9333.
Martinez JM, Afshari CA, Bushel PR, Masuda A, Takahashi T, Walker NJ: Differential toxicogenomic responses to 2,3,7,8-tetrachlorodibenzo-p-dioxin in malignant and nonmalignant human airway epithelial cells. Toxicol Sci. 2002, 69: 409-423. 10.1093/toxsci/69.2.409.
Murphy KA, Villano CM, Dorn R, White LA: Interaction between the aryl hydrocarbon receptor and retinoic acid pathways increases matrix metalloproteinase-1 expression in keratinocytes. J Biol Chem. 2004, 279: 25284-25293. 10.1074/jbc.M402168200.
Villano CM, Murphy KA, Akintobi A, White LA: 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) induces matrix metalloproteinase (MMP) expression and invasion in A2058 melanoma cells. Toxicol Appl Pharmacol. 2006, 210: 212-224. 10.1016/j.taap.2005.05.001.
Igarashi TM, Bruner-Tran KL, Yeaman GR, Lessey BA, Edwards DP, Eisenberg E, Osteen KG: Reduced expression of progesterone receptor-B in the endometrium of women with endometriosis and in cocultures of endometrial cells exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Fertil Steril. 2005, 84: 67-74. 10.1016/j.fertnstert.2005.01.113.
Haque M, Francis J, Sehgal I: Aryl hydrocarbon exposure induces expression of MMP-9 in human prostate cancer cell lines. Cancer Lett. 2005, 225: 159-166. 10.1016/j.canlet.2004.11.043.
Revich B, Aksel E, Ushakova T, Ivanova I, Zhuchenko N, Klyuev N, Brodsky B, Sotskov Y: Dioxin exposure and public health in Chapaevsk, Russia. Chemosphere. 2001, 43: 951-966. 10.1016/S0045-6535(00)00456-2.
ED-Genes. [http://www1.fukui-med.ac.jp/SEIKA2/ED-Genes.html]
Acknowledgements
We would like to thank Ms. Hiroko Matsuda, Ms. Junko Sakurai, Ms. Yuko Nobe, Ms. Tomoko Satake for their expert technical assistance. This work was supported by the Grant in Aid (No.15591729) from the Japanese Ministry of Education, Science, Sports and Culture.
Author information
Authors and Affiliations
Corresponding author
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Abe, Y., Sinozaki, H., Takagi, T. et al. Identification of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-inducible genes in human amniotic epithelial cells. Reprod Biol Endocrinol 4, 27 (2006). https://doi.org/10.1186/1477-7827-4-27
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1477-7827-4-27