Abstract
Background
The XRCC1 polymorphisms have been implicated in bladder cancer risk, but individually published studies show inconsistent results. The aim of our study was to clarify the effects of XRCC1 variants on bladder cancer risk.
Methods
A systematic literature search up to September 13, 2012 was carried out in PubMed, EMBASE and Wanfang databases, and the references of retrieved articles were screened. Crude odds ratios with 95% confidence intervals were used to assess the associations between XRCC1 Arg194Trp and Arg399Gln polymorphisms and bladder cancer risk. Heterogeneity and publication bias were also evaluated.
Results
A total of 14 and 18 studies were eligible for meta-analyses of Arg194Trp and Arg399Gln, respectively. Regrouping was adopted in accordance with the most probable appropriate genetic models. No obvious heterogeneity between studies was found. For overall bladder cancer, the pooled odds ratios for Arg194Trp and Arg399Gln were 1.69 (95% confidence interval: 1.25 to 2.28; P = 0.001) and 1.10 (95% confidence interval: 1.03 to 1.19; P = 0.008), respectively. After excluding the studies that were not in Hardy–Weinberg equilibrium, the estimated pooled odds ratio still did not change at all.
Conclusions
The meta-analysis results suggest that XRCC1 Arg194Trp and Arg399Gln polymorphisms may be associated with elevated bladder cancer risk.
Similar content being viewed by others
Background
Bladder cancer is an important health problem worldwide. It is the seventh most common malignancy in men and seventeenth in women [1]. An estimated 386,300 new cases and 150,200 deaths from bladder cancer occurred in 2008 worldwide [2]. However, the mechanism of bladder cancer is not completely clear and is considered to be a multifactorial process. The most established risk factors for bladder cancer include cigarette smoking, occupational exposure to arylamines and schistosomal infection [1]. These exogenous mutagens or carcinogens produce a wide range of DNA lesions, bulky DNA adducts, and DNA strand breaks. Epidemiologic evidence has shown that genetic variants at one or more loci result in reduced DNA repair capacity and an increased cancer risk [3–5].
DNA carrying essential heritable information must remain stable in order to undertake its key physiologic functions, but it is continually vulnerable to many types of endogenous and/or exogenous damage; thus, genetic alterations could accumulate and tumorigenesis may occur because of the damaged DNA. The DNA repair system plays a pivotal role in maintaining the genome integrity and stability through the reversal of DNA damage. If accumulated genetic alterations occurred in corresponding DNA repair genes, their reversal capacity could be damaged, possibly increasing the risk of cancer in carriers [6]. A large number of SNPs in common DNA repair genes have been identified [7] and confirmed to be associated with several sporadic cancers [8, 9].
X-ray repair cross-complementing group 1 (XRCC1), located on chromosome 19q13.2–13.3, with 33 kb in length, is an important component of base excision repair (BER) [10]. BER consists of a series of consecutive steps from the recognition and excision of a damaged base to the ligation of broken points, which are mainly conducted by XRCC1. When damage occurs, XRCC1 recruited by DNA glycosylases, acts as a platform by regulating and coordinating a whole list of BER proteins and single strand break repair (SSBR) machinery [11, 12]. Although there are more than 300 validated SNPs in the XRCC1 gene reported in the dbSNP database (http://www.ncbi.nlm.nih.gov/SNP), two genetic changes including Arg194Trp on exon 6 (rs1799782 in dbSNP, C/T) and Arg399Gln on exon 10 (rs25487 in dbSNP, G/A) are the most extensively studied. Many previous studies have been conducted to evaluate the associations of XRCC1 polymorphisms with bladder cancer risk. However, the results of these studies remain inconsistent and contradictory, partially because a single study may be too underpowered to detect a possible small effect of the polymorphism on bladder cancer, especially when the sample size is relatively small. Thus, to clarify the effect of XRCC1 variants (Arg194Trp and Arg399Gln) on bladder cancer risk, we performed a meta-analysis of all eligible studies.
Methods
Publication search
We carried out a systematic literature search in EMBASE, PubMed and Wanfang databases, covering all the papers published from their inception to September 13, 2012, using the following key words: (XRCC1 or X-ray repair cross-complementation group 1) and (bladder cancer or bladder neoplasm or bladder tumor or urothelial cancer or urinary tract cancer) and (polymorphism or variation or variant or mutation or genotype or gene). There was no language restriction. We evaluated potentially relevant papers by checking their titles and abstracts and all the studies matching the eligible criteria were retrieved. Additional studies were identified by a manual search of the references from retrieved articles and reviews.
Inclusion criteria
Studies included in the present meta-analysis had to meet all the following criteria: (a) evaluation of the XRCC1 Arg194Trp and Arg399Gln polymorphisms and the risk of bladder cancer, (b) had a case–control design or nested case–control design, (c) had sufficient data for calculating an odds ratio (OR) with 95% confidence interval (CI). If multiple publications from the same population were available, the most recent or largest study was eligible for inclusion in this meta-analysis.
Data extraction
Data were extracted independently by two authors using a predefined data collection form, with disagreements being resolved by consensus. For each study, the following information was collected: first author’s last name, publication year, the country in which the study was carried out, ethnicity, numbers of cases and controls, genotyping methods, genotypes, and allele frequency information.
Quality assessment
The quality of each study was independently appraised by the same two authors using the quality assessment criteria, which were modified on the basis of previously published meta-analysis of molecular association studies [13, 14]. The criteria consist of seven parameters of quality: representativeness of the cases, representativeness of the controls, ascertainment of bladder cancers, control selection, genotyping examination, Hardy–Weinberg equilibrium HWE) and total sample size. (The criteria are described in detail in Additional file 1: Table S1). Scores ranged from 0 (worst) to 15 (best). Studies scoring <9 were classified as low quality, and those ≥9 as high quality. Disagreements were resolved by a joint reevaluation of the original article with a third investigator.
Statistical methods
HWE in cases and controls was examined again in our meta-analysis using the goodness-of-fit test (significant at the 0.05 level). The ORs and their 95% CIs were used to calculate and assess the strength of the association between XRCC1 Arg194Trp and Arg399Gln polymorphism and the risk of bladder cancer. If there was a statistical heterogeneity among studies, the combined ORs and 95% CIs were estimated by the DerSimonian and Laird method [15] in a random-effect model. Otherwise, the ORs were obtained by the Mantel–Haenszel method [16] in a fixed effect model.
ORs 1, 2, and 3 (OR1, OR2, and OR3) were calculated for the genotypes: 1) TT versus CC, 2) CT versus CC, and 3) TT versus CT for Arg194Trp; and 1) AA versus GG, 2) GA versus GG, and 3) AA versus GA for Arg399Gln, respectively. These pairwise differences were used to determine the most appropriate genetic model. If OR1 = OR3 ≠1 and OR2 = 1, a recessive model is implied. If OR1 = OR2 ≠ 1 and OR3 = 1, a dominant model is suggested. If OR2 =1/OR3 ≠ 1 and OR1 = 1, then a complete overdominant model is indicated. If OR1 > OR2 >1 and OR1 > OR3 >1, or if OR1 < OR2 <1 and OR1 < OR3 < , then a codominant model is suggested.
Homogeneity of ORs across studies was tested by a Chi-square-based Q statistic and the I2 score. Heterogeneity was considered significant if the P-value was <0.10. The value of I2 was used to assess the degree of heterogeneity (I2 <25% no heterogeneity; I2 = 25% to 50% moderate heterogeneity; I2 >50% large or extreme heterogeneity).
Sensitivity analysis was performed in which the meta-analysis estimates were computed after omission of every study in turn. Cumulative meta-analyses of associations for each SNP were also conducted through assortment of studies with publication time.
Evaluation of publication bias
Publication bias was assessed using Begg’s test (rank correlation method) [17] and Egger’s test (linear regression method) [18]. P <0.05 was considered to be representative of a significant statistical publication bias. All of the statistical analyses were performed with STATA 11.0 (StataCorp, College Station, TX), using two-sided P-values.
Results
Characteristics of all included studies
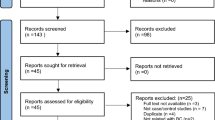
Twenty studies were included in this meta-analysis on the associations of the XRCC1 genetic polymorphisms with the risk of bladder cancer. Of the selected studies, 14 [19–32] were preliminarily appropriate for meta-analysis of the associations with bladder cancer regarding Arg194Trp, and 18 [19–22, 24–28, 30–38] were relevant to the association with Arg399Gln. Tables 1 and 2 present the basic characteristics of each study included in our meta-analysis and the corresponding genotype distributions among cases and controls. The literature search and study selection procedures are shown in Figure 1.
Quantitative synthesis
For the Arg194Trp SNP, OR1, OR2, and OR3 were 1.835 (95%CI: 1.343 to 2.507), 1.026 (95%CI: 0.920 to 1.146), and 1.581 (95%CI: 1.154 to 2.165), respectively, suggesting a recessive effect of the putative susceptibility allele T. Thus, the original grouping was collapsed, and CC and CT were combined, in accordance with a recessive model, into a C carrier group, the latter of which was compared with the TT genotype group.
For the Arg399Gln SNP, OR1, OR2, and OR3 were 0.958 (95%CI: 0.850 to 1.080), 1.095 (95%CI: 1.014 to 1.183), and 0.884 (95%CI: 0.785 to 0.997), respectively, indicating that a complete overdominant model was applicable, that is, heterozygotes are at higher risk of bladder cancer than either homozygotes (GG or AA).
As shown in Figures 2 and Table 3, the XRCC1 Arg194Trp polymorphism was associated with an increased risk for bladder cancer in all subjects (OR = 1.69, 95% CI = 1.25 to 2.28, P = 0.001). Similarly, the Arg399Gln polymorphism was also found to be significantly associated with increased risk of bladder cancer (OR = 1.10, 95% CI = 1.03 to 1.19, P = 0.008).
Odds ratios (ORs) for associations between two single nucleotide polymorphisms (Arg194Trp and Arg399Gln) in the x-ray repair cross-complementing group 1 gene ( XRCC1 ) and bladder cancer risk. The size of the black square corresponding to each study is proportional to the sample size and the center of each square represents the OR. The horizontal line shows the corresponding 95% CI of the OR. Pooled OR is represented by a hollow diamond. A) Arg194Trp TT genotypes versus the CC-plus-CT genotype; B) Arg399Gln GA genotypes versus the GG-plus-AA genotype. CI, confidence interval.
Evaluation of heterogeneity
For the Arg399Gln polymorphism, most I2 values of heterogeneity were 0% and all P values were more than 0.10, indicating no statistically significant heterogeneity between studies (Table 3). Similarly, for the Arg194Trp polymorphism, there was also no obvious heterogeneity between studies.
Sensitivity analysis
In the sensitivity analysis, the influence of each study on the pooled OR was examined by repeating the meta-analysis while omitting each study, one at a time. This procedure proved that our results were reliable and robust. In addition, when excluding the studies that were not in HWE, the estimated pooled OR still did not change at all (data not shown).
Cumulative meta-analysis
Cumulative meta-analyses of the two associations were also conducted via the assortment of studies by publication time. The 95% confidence intervals became increasingly narrower with increasing sample size, indicating that the precision of the estimates was progressively boosted by the continual addition of more cases (data not shown).
Publication bias
There was no evidence of significant publication bias either with the Begg’s test (Figures 3, P = 0.640 for Arg194Trp; P = 0.820 for Arg399Gln) or with Egger’s test (P = 0.345 for Arg194Trp; P = 0.248 for Arg399Gln).
Discussion
The Arg194Trp and Arg399Gln polymorphisms are the most well characterized XRCC1 polymorphisms, but the reported associations with bladder cancer risk among studies are inconsistent. Our present meta-analysis incorporating 20 case–control studies suggests that the Arg194Trp and Arg399Gln polymorphisms are significantly associated with increased bladder cancer risk.
In this meta-analysis, publication bias was not observed. And there was no obvious heterogeneity between studies. In addition, when repeating the meta-analysis by omitting each study, one at a time, the estimated pooled OR still did not change at all. In view of these findings, we are convinced that the results of our meta-analysis, in essence, are sound and reliable.
The results of the present study are in contrast with a previous meta-analysis published in 2008 [39], which concluded that there was no association between the XRCC1 polymorphisms and the risk for bladder cancer. However, this study only included ten studies with limited sample size (3,749 cases and 3,947 controls) and, thus, it may lack sufficient statistical power to detect the real association and may have generated a fluctuated risk estimate.
Our findings have some biological plausibility. It is widely accepted that certain genetic variants associated with repair of DNA substantially increase the risk of cancer in carriers because of the alteration of BER functions [40]. BER is the primary DNA damage repair pathway for the repairing of small base lesions resulting from oxidation and alkylation damage [41]. As one of the most important proteins in BER, XRCC1 is closely related to BER pathway coordination by interacting with most members of the BER short-patch pathway. SNP of XRCC1 may increase the risk of some types of cancer by damaging the interaction of XRCC1 with other enzymatic proteins and, consequently, altering DNA repair activity [42]; this may result in carcinogenesis, including a higher incidence of bladder cancer. Similar to the results of our study, XRCC1 polymorphisms are also reported to be associated with some other cancers. The previous three meta-analyses have confirmed that the Arg399Gln polymorphism is associated with risk of childhood acute lymphoblastic leukemia [43], breast cancer [44], and prostate cancer among Asians [45]. Dai et al. reported that the XRCC1 Arg194Trp polymorphism is associated with an increased lung cancer risk [46] and the study conducted by Li et al. suggested that the Arg194Trp polymorphism may be associated with cervical cancer risk [47]. By contrast, in our study, the Arg194Trp polymorphism was associated with disease risk only in Asians, but not in Caucasians. This is mainly because the number of Caucasians is four-fold higher than that of Asians and, therefore, the power to detect association is higher.
Several limitations of this meta-analysis should be mentioned. First, the eligibility criteria for the inclusion of subjects and sources of controls were different from each other. No guarantee could be made among all those eligible studies that there were no potential bladder cancer cases in the controls. Second, because of the lack of the individual original data, our results were just based on unadjusted estimates, and gene–gene and gene–environmental interactions were not addressed in this meta-analysis. Third, although the Begg’s test and Egger’s test did not reveal any evidence of obvious publication bias, some inevitable publication bias may exist, because only studies published in English and Chinese were included in our meta-analysis. Finally, as shown in Table 3, a borderline conclusion (OR: 1.08 (1.00 to 1.18)) of the Arg399Gln section was drawn when two studies without HWE were excluded. This conclusion actually owed much to one study [26] with a relatively large population weight, which implies the need for more well-designed studies in future.
Conclusions
In conclusion, despite some limitations, the results of our meta-analysis suggest that two polymorphisms in XRCC1 (Arg194Trp and Arg399Gln) may contribute to bladder cancer development. Whether it could be applied to genotyping for clinical assessment requires large-scale population studies among different ethnicities and regions.
Abbreviations
- BER:
-
base excision repair
- CI:
-
confidence interval
- HWE:
-
Hardy–Weinberg equilibrium
- OR:
-
odds ratio
- SNP:
-
single nucleotide polymorphism
- XRCC1:
-
X-ray repair cross-complementing group 1.
References
Murta-Nascimento C, Schmitz-Drager BJ, Zeegers MP, Steineck G, Kogevinas M, Real FX, Malats N: Epidemiology of urinary bladder cancer: from tumor development to patient’s death. World J Urol. 2007, 25: 285-295. 10.1007/s00345-007-0168-5.
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D: Global cancer statistics. CA Cancer J Clin. 2011, 61: 69-90. 10.3322/caac.20107.
Helzlsouer KJ, Harris EL, Parshad R, Perry HR, Price FM, Sanford KK: DNA repair proficiency: potential susceptiblity factor for breast cancer. J Natl Cancer Inst. 1996, 88: 754-755. 10.1093/jnci/88.11.754.
Wei Q, Spitz MR: The role of DNA repair capacity in susceptibility to lung cancer: a review. Cancer Metastasis Rev. 1997, 16: 295-307. 10.1023/A:1005852211430.
Sturgis EM, Castillo EJ, Li L, Zheng R, Eicher SA, Clayman GL, Strom SS, Spitz MR, Wei Q: Polymorphisms of DNA repair gene XRCC1 in squamous cell carcinoma of the head and neck. Carcinogenesis. 1999, 20: 2125-2129. 10.1093/carcin/20.11.2125.
Goode EL, Ulrich CM, Potter JD: Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002, 11: 1513-1530.
Miller MC, Mohrenweiser HW, Bell DA: Genetic variability in susceptibility and response to toxicants. Toxicol Lett. 2001, 120: 269-280. 10.1016/S0378-4274(01)00279-X.
Alberg AJ, Jorgensen TJ, Ruczinski I, Wheless L, Shugart YY, Berthier-Schaad Y, Kessing B, Hoffman-Bolton J, Helzlsouer KJ, Kao WH, Francis L, Alani RM, Smith MW, Strickland PT: DNA repair gene variants in relation to overall cancer risk: a population-based study. Carcinogenesis. 2013, 34: 86-92. 10.1093/carcin/bgs304.
Kury S, Buecher B, Robiou-du-Pont S, Scoul C, Colman H, Le Neel T, Le Houerou C, Faroux R, Ollivry J, Lafraise B, Chupin LD, Sébille V, Bézieau S: Low-penetrance alleles predisposing to sporadic colorectal cancers: a French case-controlled genetic association study. BMC Cancer. 2008, 8: 326-10.1186/1471-2407-8-326.
Chou WC, Wang HC, Wong FH, Ding SL, Wu PE, Shieh SY, Shen CY: Chk2-dependent phosphorylation of XRCC1 in the DNA damage response promotes base excision repair. EMBO J. 2008, 27: 3140-3150. 10.1038/emboj.2008.229.
Campalans A, Marsin S, Nakabeppu Y, O’Connor TR, Boiteux S, Radicella JP: XRCC1 interactions with multiple DNA glycosylases: a model for its recruitment to base excision repair. DNA Repair (Amst). 2005, 4: 826-835. 10.1016/j.dnarep.2005.04.014.
Marsin S, Vidal AE, Sossou M, Menissier-de Murcia J, Le Page F, Boiteux S, de Murcia G, Radicella JP: Role of XRCC1 in the coordination and stimulation of oxidative DNA damage repair initiated by the DNA glycosylase hOGG1. J Biol Chem. 2003, 278: 44068-44074. 10.1074/jbc.M306160200.
Thakkinstian A, McKay GJ, McEvoy M, Chakravarthy U, Chakrabarti S, Silvestri G, Kaur I, Li X, Attia J: Systematic review and meta-analysis of the association between complement component 3 and age-related macular degeneration: a HuGE review and meta-analysis. Am J Epidemiol. 2011, 173: 1365-1379. 10.1093/aje/kwr025.
Yu H, Liu H, Wang LE, Wei Q: A functional NQO1 609C>T polymorphism and risk of gastrointestinal cancers: a meta-analysis. PLoS One. 2012, 7: e30566-10.1371/journal.pone.0030566.
DerSimonian R, Laird N: Meta-analysis in clinical trials. Control Clin Trials. 1986, 7: 177-188. 10.1016/0197-2456(86)90046-2.
Mantel N, Haenszel W: Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959, 22: 719-748.
Begg CB, Mazumdar M: Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994, 50: 1088-1101. 10.2307/2533446.
Egger M, Davey Smith G, Schneider M, Minder C: Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997, 315: 629-634. 10.1136/bmj.315.7109.629.
Stern MC, Umbach DM, van Gils CH, Lunn RM, Taylor JA: DNA repair gene XRCC1 polymorphisms, smoking, and bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2001, 10: 125-131.
Wu W: Study on the Relationship between polymorphisms of JWA and XRCCl Genes and the Susceptibility to Bladder Cancer [D]. 2005, Lanzhou: Lanzhou University, (Chinese)
Matullo G, Dunning AM, Guarrera S, Baynes C, Polidoro S, Garte S, Autrup H, Malaveille C, Peluso M, Airoldi L, Veglia F, Gormally E, Hoek G, Krzyzanowski M, Overvad K, Raaschou-Nielsen O, Clavel-Chapelon F, Linseisen J, Boeing H, Trichopoulou A, Palli D, Krogh V, Tumino R, Panico S, Bueno-De-Mesquita HB, Peeters PH, Lund E, Pera G, Martinez C, Dorronsoro M: DNA repair polymorphisms and cancer risk in non-smokers in a cohort study. Carcinogenesis. 2006, 27: 997-1007.
Wu X, Gu J, Grossman HB, Amos CI, Etzel C, Huang M, Zhang Q, Millikan RE, Lerner S, Dinney CP, Spitz MR: Bladder cancer predisposition: a multigenic approach to DNA-repair and cell-cycle-control genes. Am J Hum Genet. 2006, 78: 464-479. 10.1086/500848.
Zhang W, Xiang YB, Cheng JR, Shao CX, Fang RR, Yuan JM, Gao YT: A case–control study of polymorphism of XRCC1 gene and the risk of bladder cancer. China Cancer. 2006, 15: 667-672. In Chinese
Sak SC, Barrett JH, Paul AB, Bishop DT, Kiltie AE: DNA repair gene XRCC1 polymorphisms and bladder cancer risk. BMC Genet. 2007, 8: 13-
Figueroa JD, Malats N, Real FX, Silverman D, Kogevinas M, Chanock S, Welch R, Dosemeci M, Tardon A, Serra C, Carrato A, García-Closas R, Castaño-Vinyals G, Rothman N, García-Closas M: Genetic variation in the base excision repair pathway and bladder cancer risk. Hum Genet. 2007, 121: 233-242. 10.1007/s00439-006-0294-y.
Andrew AS, Karagas MR, Nelson HH, Guarrera S, Polidoro S, Gamberini S, Sacerdote C, Moore JH, Kelsey KT, Demidenko E, Vineis P, Matullo G: DNA repair polymorphisms modify bladder cancer risk: a multi-factor analytic strategy. Hum Hered. 2008, 65: 105-118. 10.1159/000108942.
Hsu LI, Chiu AW, Huan SK, Chen CL, Wang YH, Hsieh FI, Chou WL, Wang LH, Chen CJ: SNPs of GSTM1, T1, P1, epoxide hydrolase and DNA repair enzyme XRCC1 and risk of urinary transitional cell carcinoma in southwestern Taiwan. Toxicol Appl Pharmacol. 2008, 228: 144-155. 10.1016/j.taap.2007.12.003.
Fontana L, Bosviel R, Delort L, Guy L, Chalabi N, Kwiatkowski F, Satih S, Rabiau N, Boiteux JP, Chamoux A, Bignon YJ, Bernard-Gallon DJ: DNA repair gene ERCC2, XPC, XRCC1, XRCC3 polymorphisms and associations with bladder cancer risk in a French cohort. Anticancer Res. 2008, 28: 1853-1856.
Narter KF, Ergen A, Agachan B, Gcjrmus U, Timirci O, Isbir T: Bladder cancer and polymorphisms of DNA repair genes (XRCC1, XRCC3, XPD, XPG, APE1, hOGGl). Anticancer Res. 2009, 29: 1389-1394.
Wang M, Qin C, Zhu J, Yuan L, Fu G, Zhang Z, Yin C: Genetic variants of XRCC1, APE1, and ADPRT genes and risk of bladder cancer. DNA Cell Biol. 2010, 29: 303-311. 10.1089/dna.2009.0969.
Bianchino G, Cittadini A, Grieco V, Traficante A, Zupa A, Improta G, Aieta M, Sgambato A: Polymorphisms of the CYP1A1, CYP2E1 and XRCC1 genes and cancer risk in a Southern Italian population: a case–control study. Anticancer Res. 2011, 31: 1359-1365.
Mittal RD, Mandal RK, Gangwar R: Base excision repair pathway genes polymorphism in prostate and bladder cancer risk in North Indian population. Mech Ageing Dev. 2012, 133: 127-132. 10.1016/j.mad.2011.10.002.
Shen M, Hung RJ, Brennan P, Malaveille C, Donato F, Placidi D, Carta A, Hautefeuille A, Boffetta P, Porru S: Polymorphisms of the DNA repair genes XRCC1, XRCC3, XPD, interaction with environmental exposures, and bladder cancer risk in a case–control study in northern Italy. Cancer Epidemiol Biomarkers Prev. 2003, 12: 1234-1240.
Sanyal S, Festa F, Sakano S, Zhang Z, Steineck G, Norming U, Wijkstrom H, Larsson P, Kumar R, Hemminki K: Polymorphisms in DNA repair and metabolic genes in bladder cancer. Carcinogenesis. 2004, 25: 729-734.
Broberg K, Bjork J, Paulsson K, Hoglund M, Albin M: Constitutional short telomeres are strong genetic susceptibility markers for bladder cancer. Carcinogenesis. 2005, 26: 1263-1271.
Karahalil B, Kocabas NA, Ozcelik T: DNA repair gene polymorphisms and bladder cancer susceptibility in a Turkish population. Anticancer Res. 2006, 26: 4955-4958.
Arizono K, Osada Y, Kuroda Y: DNA repair gene hOGG1 Codon 326 and XRCC1 Codon 399 polymorphisms and bladder cancer risk in a Japanese population. Jpn J Clin Oncol. 2008, 38: 186-191. 10.1093/jjco/hym176.
Zhi Y, Yu J, Liu Y, Wei Q, Yuan F, Zhou X, Song B, Chen Z, Yang J: Interaction between polymorphisms of DNA repair genes significantly modulated bladder cancer risk. Int J Med Sci. 2012, 9: 498-505. 10.7150/ijms.4799.
Wang C, Sun Y, Han R: XRCC1 genetic polymorphisms and bladder cancer susceptibility: a meta-analysis. Urology. 2008, 72: 869-872. 10.1016/j.urology.2007.12.059.
Monaco R, Rosal R, Dolan MA, Pincus MR, Brandt-Rauf PW: Conformational effects of a common codon 399 polymorphism on the BRCT1 domain of the XRCC1 protein. Protein J. 2007, 26: 541-546. 10.1007/s10930-007-9095-y.
Maynard S, Schurman SH, Harboe C, de Souza-Pinto NC, Bohr VA: Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009, 30: 2-10.
Tudek B: Base excision repair modulation as a risk factor for human cancers. Mol Aspects Med. 2007, 28: 258-275. 10.1016/j.mam.2007.05.003.
Wang L, Yin F, Xu X, Hu X, Zhao D: X-ray repair cross-complementing group 1 (XRCC1) genetic polymorphisms and risk of childhood acute lymphoblastic leukemia: a meta-analysis. PLoS One. 2012, 7: e34897-10.1371/journal.pone.0034897.
Wu K, Su D, Lin K, Luo J, Au WW: XRCC1 Arg399Gln gene polymorphism and breast cancer risk: a meta-analysis based on case–control studies. Asian Pac J Cancer Prev. 2011, 12: 2237-2243.
Wei B, Zhou Y, Xu Z, Ruan J, Zhu M, Jin K, Zhou D, Hu Q, Wang Q, Wang Z, Yan Z: XRCC1 Arg399Gln and Arg194Trp polymorphisms in prostate cancer risk: a meta-analysis. Prostate Cancer Prostatic Dis. 2011, 14: 225-231. 10.1038/pcan.2011.26.
Dai L, Duan F, Wang P, Song C, Wang K, Zhang J: XRCC1 gene polymorphisms and lung cancer susceptibility: a meta-analysis of 44 case–control studies. Mol Biol Rep. 2012, 39: 9535-9547. 10.1007/s11033-012-1818-2.
Li Y, Liu F, Tan SQ, Wang Y, Li SW: X-ray repair cross-complementing group 1 (XRCC1) genetic polymorphisms and cervical cancer risk: a HuGE systematic review and meta-analysis. PLoS One. 2012, 7: e44441-10.1371/journal.pone.0044441.
Acknowledgements
This study was supported by grants from the National Key Clinical Specialty Construction Project of China, Key medical disciplines of Zhejiang province, Combination of traditional Chinese and Western medicine key disciplines of Zhejiang Province (2012-XK-A23), Health sector scientific research special project (201002010), National Natural Science Foundation of China (Grant No. 30900552) and Zhejiang Provincial Natural Science Foundation of China (Z2090356).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LPX, YQM and XX developed the study concept and participated in its design, data extraction, statistical analysis, manuscript drafting and editing. YWL and HC participated in the literature research, manuscript drafting and editing. JW and ZHH participated in design and data extraction. XLX and YZ participated in manuscript drafting, editing and statistical analysis. All authors read and approved the final manuscript.
Yeqing Mao, Xin Xu contributed equally to this work.
Electronic supplementary material
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Mao, Y., Xu, X., Lin, Y. et al. Quantitative assessment of the associations between XRCC1 polymorphisms and bladder cancer risk. World J Surg Onc 11, 58 (2013). https://doi.org/10.1186/1477-7819-11-58
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1477-7819-11-58