Abstract
Background
Osteoarthritis (OA) is a destructive joint disease and there are no known biomarkers available for an early diagnosis. To identify potential disease biomarkers and gain further insight into the disease mechanisms of OA we applied quantitative proteomics with SILAC technology on the secretomes from chondrocytes of OA knees, designated as high Mankin (HM) scored secretome. A quantitative comparison was made between the secretomes of the medial and lateral femur condyle chondrocytes in the same knee since the medial femur condyle is usually more affected in OA than the lateral condyle, which was confirmed by Mankin scoring. The medial/lateral comparison was also made on the secretomes from chondrocytes taken from one individual with no clinically apparent joint-disease, designated as low Mankin (LM) scored secretome.
Results
We identified 825 proteins in the HM secretome and 69 of these showed differential expression when comparing the medial and lateral femoral compartment. The LM scored femoral condyle showed early signs of OA in the medial compartment as assessed by Mankin score. We here report the identification and relative quantification of several proteins of interest for the OA disease mechanism e.g. CYTL1, DMD and STAB1 together with putative early disease markers e.g. TIMP1, PPP2CA and B2M.
Conclusions
The present study reveals differences in protein abundance between medial/lateral femur condyles in OA patients. These regulatory differences expand the knowledge regarding OA disease markers and mechanisms.
Similar content being viewed by others
Introduction
Osteoarthritis (OA) is a complex age-related polygenetic progressive degenerative disease of the synovial joint characterized by gradual destruction of the articular cartilage and subchondral bone leading to a loss of joint function. The gradual destruction of the articular cartilage causes joint pain and stiffness, which slowly disables the patient. The OA disease could affect a single joint, several joints or be more generalized. The disease prevalence is higher in women [1–4] and when the knee joint is affected in OA there is a predominant tissue degradation of the medial compartment as compared to the lateral compartment [1, 2, 5]. Due to the lack of disease modifying treatments the only treatment option for the degenerated joint is eventually a total joint replacement. However, isolated moderate to large knee cartilage defects due to trauma benefit from autologous chondrocytes transplantation procedures [6, 7].
The etiology of OA is still elusive although many studies have been performed in order to find pathological mediators and biomarkers [8–11]. When searching for disease processes in OA the whole joint should be considered a diseased organ where the synovium, articular chondrocytes as well as the patellar fat pad contribute with secreted factors, which may have catabolic and/or anabolic effects on articular cartilage [12–15]. Inflammation has been proposed to cause catabolic processes in the joint resulting in cartilage erosion, see Abramson and Attur [16] for a comprehensive review. Signaling pathways as well as autocrine and paracrine mediators have been studied in OA in order to reveal the processes behind the slow regeneration of cartilage [17] and to identify putative drug targets [18]. There have been several genome-wide association studies of osteoarthritis and one of the largest is the arcOGEN study that in its first report from a genome-wide association scan for knee and hip osteoarthritis in 3,177 cases and 4,894 population-based controls was unable to find a genetic linkage to the disease. The conclusion from the study was that “data suggests that osteoarthritis is a highly polygenic disease with multiple risk variants conferring small effects” and that additional larger populations studies are needed [19]. In a second report five genome-wide significant foci were identified [20]. When searching the OMIM database for genes connected to OA GDF5 and FRZB among others are considered potential candidates.
The introduction of global proteomic techniques enables studies of the functional proteome after posttranslational modifications and protein processing, which give a more comprehensive picture in the evaluation of disease hypothesis than solely gene expression analysis. Stable isotope labeling by amino acids in cell culture (SILAC) is a metabolic labeling technique that incorporates nonradioactive isotope labeled amino acids in all newly synthesized proteins [21]. The isotope labeled proteome from one cell culture can be qualitatively and quantitatively compared to another culture condition or cell line cultured with normal amino acids using mass spectrometry (MS). Hence, SILAC combined with MS makes a powerful tool capable of analyzing global proteomic variances. Cultured chondrocytes from OA patients have been studied [22–24] in functional and comparative studies and SILAC approaches in cartilage explants have been tried out but failed to label the total proteome [22].
Our primary hypothesis was that the secretome from culture expanded OA chondrocytes is relevant when compared to the cartilage explants secretome. Secondly we hypothesized that since in OA the medial compartment of the knee joint is more often and more severely affected as compared to the lateral compartment, differences should exist in the secretomes since the cells originate from cartilage with different grades of degeneration. Further, the potential protein abundance differences could expand the knowledge regarding OA disease markers and mechanisms. We here demonstrate that the SILAC technology enables a unique bilateral comparison in the same patient with new interesting results and to our knowledge this is the first time such a comparison has been done.
Results
Characterization of cartilage biopsies for monolayer cultures
Total knee replacement biopsies from five OA patients, included in the monolayer culture, were characterized by Mankin score and designated high Mankin scored (HM) patients. The characterization resulted in an average Mankin score for the femoral medial condyle of 9.8 and for the femoral lateral condyle of 4.9 (Table 1). The same characterization was done for the macroscopically healthy cartilage from one individual, which received Mankin score 2.7 for the medial femoral condyle and 0.3 for the lateral femoral condyle (Table 1) and was subsequently designated as a low Mankin score (LM) patient although the medial condyle was slightly affected by an early OA.
The relevance of monolayer culture as an experimental model to study the chondrocyte secretome
A previous report has demonstrated the difficulty to incorporate stable isotope labels into proteins in cartilage explant cultures with a relatively small number of identified labeled proteins as a result [22]. This is in agreement with our own results from SILAC labeling of OA explants where stable isotope labeled proteins were present at very low abundance and thus not useful for quantitative analysis (data not shown). We therefore performed stable isotope labeling with amino acids in cell culture (SILAC) of chondrocytes in monolayer cultures. However, in order to evaluate the monolayer culture as an experimental model relevant for studies of differences in the secretome of medial femur condyle and lateral femur condyle positions of the OA knee joint we compared the secretomes in the OA explants and monolayer cultures. For this specific comparison we used the combined secretomes identified from the five monolayer cultures isolated from OA cartilage with high Mankin score and the combined secretomes from the two OA cartilage explants. In the supernatants of OA cartilage explant cultures we identified a total of 344 protein groups at a false discovery rate (FDR) of 1% (Additional file 1: Table S1). An additional filtering for proteins identified with at least two peptides was made, which yielded 320 protein groups. Monolayer cultures of femoral chondrocytes from the five high Mankin scored individuals were grown to full labeling and conditioned with serum-free media for 24 hours. Analysis of the supernatants from the HM monolayer cultures identified a total of 825 protein groups at a FDR of 1% (Additional file 2: Table S2) and additional filtering for proteins identified with at least two peptides yielded 727 protein groups. A comparison of the identified proteins from cartilage explants and monolayer cultured chondrocytes filtered for 1% FDR and identified with at least two peptides showed that 172 proteins were identified in both groups, which represented 54% of the proteins identified in the explant secretome (Additional file 3: Table S3). Polacek et al. used chondrocytes from autologous chondrocyte implantations (ACI) to establish monolayer cultures. Out of the 100 unique proteins identified from their analysis 84 overlapped with our study.
Among the proteins identified in both explant and monolayer cultures were several cartilage extracellular matrix remodeling (ECM) proteins, e.g. cartilage oligomeric matrix protein (COMP), matrix metalloproteinase (MMP)-1, -2, -3 and -10, metalloproteinase inhibitor (TIMP) 1 and 2 and several isoforms of collagen. Genetic association for disease analysis of the 172 proteins common for both monolayer chondrocytes and explant cultures showed the term osteoarthritis to be highly connected to the protein list (P-value ≤ 0.01). Gene ontology analysis for biological process of the 172 common proteins also showed the most enriched terms to be involved in ECM organization and inflammation processes further supporting the hypothesis that the monolayer cultured OA chondrocytes could partly represent the OA secretome (Additional file 4: Table S4, sheet 1). However the well-known loss of phenotype of chondrocytes when subjected to monolayer cultures was also demonstrated by presence of Collagen type I alpha 1 and 2 subunits and absence of Collagen type II subunits in the monolayer secretome (Additional file 2: Table S2).
Secretome profile of high and low Mankin scored femur chondrocytes
The proteomic analysis of the chondrocyte secretomes from five OA patients undertaking a total knee replacement operation resulted in a comprehensive secretome profile of femoral condyle knee OA chondrocytes. In total, 825 protein groups were identified with a 1% FDR (Additional file 2: Table S2). The corresponding analysis of a low Mankin scored individual resulted in 528 identified protein groups (Additional file 5: Table S5). Gene ontology enrichment analysis of the high Mankin scored secretome regarding cellular compartment and biological process using the DAVID bioinformatics resource showed the highest statistical significance for the GO terms GO:0044421 extracellular region part and GO:0030198 extracellular matrix organization (Additional file 4: Table S4, sheet 2–3). The corresponding analysis of the low Mankin scored secretome showed high statistical significance for the GO term GO:0044421 extracellular region part and interestingly high statistical significance for the biological process terms GO:0030199 collagen fibril organization and GO:0006916 anti-apoptosis (Additional file 4: Table S4, sheet 4–5).
Differentially expressed proteins in the femur medial and lateral knee compartments in OA patients
The secretome analysis from femoral condyle medial and lateral high Mankin chondrocytes within the same knee of five OA patients showed significant differences in the protein amounts of 69 proteins, (Table 2 and Additional file 6: Table S6), among these, 28 protein groups were medially abundant and 41 were laterally abundant (Figure 1). Six proteins were significantly regulated in at least three out of five patients, when comparing the medial and lateral positions. Gene ontology biological process term enrichment analysis of the significantly regulated high Mankin scored secretome showed the highest statistically significant enrichment of the terms GO:0006954 inflammatory response and GO:0009611 response to wounding (Additional file 4: Table S4, sheet 6). Proteins involved in these processes were Transferrin, Stabilin1, Insulin, Clusterin, S100 calcium binding protein A9, Annexin A1, Desmoplakin, Enolase 3, Complement component 1, r subcomponent, Insulin-like growth factor binding protein 4 and Macrophage migration inhibitory factor. Cellular compartment gene ontology analysis also showed the highest significance for GO:0005576 extracellular region (Additional file 4: Table S4, sheet 7).
Differentially expressed proteins in the femur medial and lateral knee compartments in a low Mankin scored patient
Histograms of Log2 values of the protein Heavy/Light (H/L) ratios for the low Mankin scored individual were produced, both labeling experiments resulted in a bimodal distribution (Figure 2). The Significance B could not be calculated since the underlying assumption in this calculation is normal distribution of the data. For these samples the Log2 values were normalized to the most frequent value and proteins with a fold-change above two in either of the experiments were selected. This resulted in 200 identified proteins in the low Mankin scored individual with different levels when comparing the femoral medial and lateral compartments. The medial femoral compartment showed a higher abundance of 34 proteins out of which several are known to affect cartilage homeostasis e.g. TIMP1, TIMP2, SPARC, Col6A1 and Col12A1. Further, Insulin Growth Factor binding protein 6, 7, 3 and 4 were also present at a higher level in the medial compartment of the femoral condyle (Table 3 and Figure 1). Gene ontology analysis of biological process showed statistically significant enrichment for the terms GO:0001501 skeletal system development, GO:0042127 regulation of cell proliferation, and GO:0030198 extracellular matrix organization (Additional file 4: Table S4, sheet 8). In addition, 166 proteins were present at a higher level within the femoral condyle lateral compartment, as compared to the medial compartment, in the low Mankin scored individual (Table 4, Additional file 7: Table S7 and Figure 1). The laterally abundant protein groups showed significant term enrichment results in gene ontology analysis for GO:0030036 actin cytoskeleton organization e.g. Destrin, GO:0006006 glucose metabolic process e.g. Enolase 1 and GO:0006916 anti-apoptosis e.g. Annexin a1 (Additional file 4: Table S4, sheet 9). Interestingly, the femoral medial and lateral protein SILAC ratios seen in the LM scored individual were lost in the HM patients, i.e. the SILAC ratio between the medial and lateral femur condyle compartments were close to one (Tables 3, 4 and Additional file 7: Table S7). Thus, the quantitative difference in protein secretion between the medial and lateral compartments found in the low Mankin scored individual is normalized in the medial and lateral compartments in the high Mankin scored individuals.
Verification of MS results by ELISA
The MS data of two of the most prominent ECM remodeling proteins, i.e. TIMP1 and MMP2, were verified with ELISA. The ELISA confirmed that the levels of MMP2 and the known MMP inhibitor TIMP1 were approximately equal between the medial and lateral compartments in the HM individuals. Further the ELISA also verified that the SILAC ratio values for the LM individual showed a medial abundance of TIMP1 (Figure 3). MMP2 was identified in the secretome from the LM individual but a SILAC ratio could not be generated. Further analysis of the MS data showed that the identified MMP2 peptides from the LM individual included the stable isotope labeled amino acids indicating that the MMP2 was mainly generated from the medial compartment of the LM femoral condyle. ELISA confirmed this observation since the lateral chondrocytes from the LM individual did not secrete any MMP2 at levels detectable by ELISA while the medial chondrocytes secreted MMP2 in amounts within the levels from the HM chondrocytes (Figure 3). Taken together ELISA confirmed the MS results for TIMP1 and MMP2 in all samples.
ELISA of MMP2 and its inhibitor TIMP1 levels in the HM individuals (A) and the LM individual (B). The two measuring points in the LML and LMM group represent the two individually labeled culturing experiments of the low Mankin scored individual. High Mankin Scored Lateral Chondrocytes (HML), High Mankin Scored Medial Chondrocytes (HMM), Low Mankin Scored Lateral Chondrocytes (LML) and Low Mankin Scored Medial Chondrocytes (LMM).
Proteins uniquely identified in low Mankin scored chondrocytes
We finally addressed the possibility of identifying unique proteins only secreted in the LM scored secretome and not in the secretome of the five HM scored individuals. Indeed, we found 25 unique proteins in the LM scored individual where 5 were found in both LM labeled experiments (EPB41L2, AKAP12, FABP3, SCRN-1, CDV3) (Additional file 8: Table S8).
Discussion
This study was designed to quantitatively compare, using SILAC technology, the secretome from medial and lateral femoral condyle chondrocytes obtained from OA patients undergoing total knee replacement and from an aged and sex matched individual with no clinical joint disease as a hypothesis generating approach towards a better understanding of OA. The LM sample became highly interesting since the LM medial side data suggested an early stage OA compared to the non-affected LM lateral side, such early stage OA samples are very rare in literature and have never been examined with the SILAC-technology before.
Results from the first studies that applied the SILAC technology on articular cartilage were qualitative identifying newly synthesized proteins [22, 25]. Later studies performed by Calamia et al. were both qualitative and quantitative [23, 24] but to our knowledge our study is the first that qualitatively and quantitatively compares medial and lateral compartments with different grades of cartilage degeneration within the same knee.
Previous attempts to label the explant secretome by the SILAC-technique has proven difficult in articular cartilage and our own attempt to label the explant secretome gave similar results (data not shown) as the study by Polacek et al. where only 25 to 30% of the explant secretome was labeled possibly due to the slow cell cycle of chondrocytes within the cartilage ECM or interference from the ECM itself [22]. This is a methodological drawback since quantitative evaluation of SILAC experiments is only interpretable if the labeled proteome is fully saturated with the stable isotope. Therefore we widened our study with a first aim to address the hypothesis whether the monolayer chondrocyte culture secretome could mirror the secretome from native explants and thus partly represent the OA secretome. The present study demonstrates that a substantial proportion of the detected proteins secreted from explants (54%) were also detected among the secreted proteins from monolayer cultures and that several proteins important for ECM remodeling and reflecting OA disease were found in the common secretome. Our conclusion is that although monolayer culture has its limitations in respect of phenotype change e.g. regarding Collagen type II the identified monolayer secretome indicate an association with OA. However, to validate the findings in the present study there is an absolute need for experimental improvements in future quantitative studies of the explant secretome. Such improvements could involve modifications of the incubation protocols and use of thinner explant slices. An alternative labeling method that could have been used is iTRAQ/TMT but it does not solve the problem with quantitative analysis of newly synthesized proteins versus proteins present within the cartilage extracellular matrix.
We studied the secreted proteins from chondrocytes as they have particular importance in tissue maintenance and regeneration as well as their putative usage as biomarkers. Further, it is important to study the secreted proteins since only approximately 7% of the proteins found in the intracellular proteome of chondrocytes may have a direct role in cartilage ECM synthesis/turnover [23].
The cartilage specimens used in this study were scored according to the Mankin scoring system [26], a well accepted and validated score for histological grading of cartilage histology [27]. The sample obtained from the patient with no history of joint disease had a medial Mankin score of 2.7 and a lateral Mankin score of 0.3. The different scores indicated that although the LM scored patient was asymptomatic from any joint disease the medial compartment was weakly affected and could hypothetically be regarded as a mild early stage of OA. The changes in protein secretion between the medial and lateral side are thus interesting as potential markers of early OA. The single LM scored sample gave no possibility to draw any statistical conclusions regarding the proteomic state within normal knee joints. However, the results from the LM scored specimen are well experimentally documented by repeated independent cultures from the medial and lateral compartments in both labeled and unlabeled medium, respectively. In this way we could analyze the secretome twice with inverted labeling conditions, which gave strong technical data that supported the validity of the results.
In the HM scored OA patients we identified 825 proteins and 69 of these showed to have a significantly changed abundance when comparing the femoral medial and lateral compartments.
The 69 significantly altered proteins were further analyzed in the DAVID where the proteins were significantly annotated to the terms inflammatory response and response to wounding, which correlates with OA disease parameters. Enolase 3, Stabilin 1 and Transferrin were among the six proteins that were significantly regulated in three of five HM individuals and interestingly annotated as response to wounding (Table 2). Enolase 3 was medially elevated and Stabilin 1 and Transferrin were laterally elevated. Interestingly, Stabilin 1 has been shown to be involved in macrophage mediated clearance of SPARC, a protein which is thought to be involved in mineralization of bone and is known to be up-regulated in OA [28–31]. In this study Stabilin 1 is less abundant in the HM femoral medial compartment, which could result in a reduced localized SPARC clearance contributing to the medial compartment OA pathogenesis. Transferrin is known to be synthesized by hypertrophic chondrocytes and is thought to be a chemotactic factor promoting migration of endothelial cells [32]. In OA cartilage this could be a reparative response that is down-regulated in the more damaged femur medial compartment. The remaining three significantly regulated proteins were not included in a single significantly enriched GO term; dystrophin (DMD); small proline rich protein (SPRR2C) and Collapsin response mediator protein 4 long variant (DPYSL3), however SPRR2C was included in the significantly enriched term GO:0009913 epidermal cell differentiation. SPRR2C and DPYSL3 have important cellular functions but no known function in cartilage [33, 34]. Interestingly, DMD deficient female mice that had the opportunity to voluntary exercise were affected by a significantly reduced cartilage thickness and cartilage tissue area on the proximal femur head compared to wild type control [35]. The present study shows DMD levels to be significantly reduced in the femoral medial more damaged compartment compared to the lateral more intact compartment of the OA knee as evaluated by Mankin score. Thus DMD may have an ECM preserving role in hyaline cartilage and interestingly DMD was annotated as GO:0042592 homeostatic process. To our knowledge this study is the first to report the involvement of DMD in human OA. Taken together, the MS results from the HM scored individuals include a comprehensive secretome of the OA phenotype together with proteins that may mark the OA progression between the affected lateral compartment to the heavily affected medial compartment.
We found several interesting proteins in the LM scored individual to be elevated with a fold change over 2 in the femoral medial compartment where Cystatin-3, Timp1 and B2M were among the most medially elevated proteins. Cystatin-3 is a cysteine protease inhibitor, which is known to modulate the effect of the cysteine proteinase Cathepsin B [36]. Cathepsin B was also elevated in the femoral medial compartment of the healthy individual, which may indicate an early state of OA in the medial compartment supported by the facts that elevated Cathepsin B levels has been found in OA specimens [37]. This is in accordance with the higher Mankin score in the femur medial compartment of the LM scored individual. The effect of Cathepsin B may however be balanced by the elevated levels of Cystatin-3.
TIMP1 and TIMP2 are known endogenous inhibitors of MMPs, which makes them key factors in the cartilage ECM homeostasis. The elevated TIMP1 and TIMP2 levels found in the LM score individual in the femoral medial chondrocyte secretome may be a reactive mechanism in response to an early OA as the medial condyle had a higher Mankin score. This was further supported by the identification of MMP2 in the MS-analysis, even though the levels in the lateral compartment were not sufficient for determination of a SILAC-ratio. Furthermore, ELISA confirmed MMP2 to be only present in the medial secretome of the LM individual. The C-terminal fragments of Procollagen C-endopeptidase enhancer 1 (PCPE) have been shown to inhibit MMP2 [38]. PCPE, also known as PCOLCE, levels were elevated in the LM femoral medial chondrocyte secretome, which is the same abundance pattern as MMP2 in this individual. PCOLCE was further expressed in all MMP2 expressing HM scored chondrocytes but was absent in the secretome from the HM individual nr 3 where no MMP2 was detected by either MS or ELISA. Taken together these results indicate that MMP2 and PCOLCE levels are connected to each other possibly due to a protective feedback loop. B2M has previously been proposed to be involved in OA pathogenesis as B2M is expressed in OA cartilage and may influence chondrocyte gene expression as well as inhibit proliferation of chondrocytes [39]. The medially up-regulated B2M levels found in the LM scored individual could potentially balance the proliferative stress signals that may be initiated by normal joint usage and/or be the early response to a low grade OA and thus a potential early marker of OA. Our group has previously reported that chondrocytes from different locations within the knee joint have different production of proteoglycans and collagens together with different chondrogenic potential in vitro [40]. Interestingly, the present study demonstrates that the differences in abundance of the medially/laterally regulated proteins in the LM scored individual were abolished in OA patients indicating that a homeostatic balance within the LM scored knee may be impaired in the HM scored OA knee, however this hypothesis must be further evaluated in the future.
The low TIMP1 and MMP2 ELISA values for the HM3 patient (with the lowest Mankin score within the HM group) could indicate an intermediate disease stage for this patient, which is further supported by the similarities in SILAC ratios with the LM scored patient. Such SILAC ratios were seen for IGFBP7, EFEMP1, FSTL1, CHI3L1, IGFBP4, SPARC and B2M. EFEMP1 or formerly Fibulin-3 was demonstrated to have a negative regulation on chondrocyte differentiation and was proposed as a diagnostic biomarker for OA [41, 42]. FSTL1 has been demonstrated to be correlated to the severity of OA disease as well as CHI3L1 [43–45]. SPARC and B2M are correlated to OA while the IGFBP7 has not been correlated to OA previously [30, 39, 46]. IGFBP4 is suggested to inhibit the canonical Wnt signaling pathway, which makes elevated levels of IGFBP4 on the medial side in the LM scored specimen an interesting finding as stimulated Wnt signaling is known to be involved in OA pathogenesis [47–50]. This SILAC ratio pattern may indicate an OA initiation in the medial femur condyle of the LM scored individual and a counteracting reparative process, which has balanced cartilage homeostasis in this aged and sex matched individual who never developed clinical signs of OA. When the lateral side becomes more affected by the OA, as in the HM3 patient, the ratio becomes smaller towards a ratio close to one seen in the highest Mankin scored patients.
One of the medially abundant proteins in the LM individual is the Cytokine like protein 1 (CYTL1) which is only found in two HM patients with an elevated level on the femur lateral side. CYTL1 is a novel autocrine regulatory factor that regulates chondrogenesis of mouse mesenchymal cells [51]. In a recent knockout study deletion of the CYTL1 gene did not affect chondrogenesis or cartilage development. However, CYTL1 knockout mice were more sensitive to osteoarthritic cartilage destruction and expression levels of CYTL1 were markedly decreased in OA cartilage of humans and experimental mice [52]. Our SILAC data thus support the hypothesis that CYTL1 is required for the maintenance of cartilage homeostasis, and loss of CYTL1 function is associated with OA cartilage destruction.
SILAC ratios showing laterally abundant proteins in the LM individual were also analyzed and showed several proteins designated as anti-apoptotic (e.g. Annexin 1 and 5). Chondrocyte derived S100A8 and S100A9 have a potential role in inflammatory arthritis in initiating early cartilage degradation by up-regulating MMPs and aggrecanases [53], partly supported by our results with laterally abundant S100A11 and medially abundant S100A8 in the LM scored individual.
Finally we analyzed the MS data for unique proteins in the LM scored secretome that were not found in any of the high Mankin scored patients. Interestingly we found 25 unique proteins in the LM scored secretome and all proteins found were elevated on the lateral femoral side except for the coatomer subunit zeta-1. Five of the proteins were also found in the inversely labeled LM scored experiment i.e. EPB41L2, AKAP12, FABP3, SCRN-1, CDV3 and are thus of high relevance and certainty. The A-kinase anchor protein 12 (AKAP12) has a pivotal role in regulation of cellular adhesion dynamics and is also a putative inhibitor of angiogenesis through down-regulation of MMP9 expression [54]. MMP9 inhibition through AKAP12 may thus suppress the typical hallmarks of OA in the LM scored individual i.e. ECM degradation and angiogenesis in the cartilage. Secernin-1 (SCRN-1) regulates exocytosis in mast cells and has recently been proposed as a prognostic marker in synovial sarcoma where high expression is associated with a better prognosis possibly due to a higher grade of differentiation [55, 56]. Thus, high SCRN-1 levels could mark a differentiated cartilage, and in the case of the LM individual mark a more dedifferentiated OA-like state in the medial compartment as the levels were medially lowered. However, these conclusions may be tissue specific as SCRN-1 expression has also been proposed as a prognostic marker in colorectal cancer where elevated expression was associated with a worse prognosis [57]. The FABP3 protein is related to regulation of apoptosis where it both promotes and inhibits apoptosis, which may be of interest in further studies regarding its role in OA [58–60]. Among proteins that only generated a protein SILAC ratio in one LM experiment PPP2CA is the catalytic subunit of protein phosphatase 2A (PPP2A). Inhibition of PPP2A in TGFbeta stimulated normal human chondrocytes has an anti apoptotic effect [61]. Inhibition of PPP2A is also suggested to have a chondrogenic effect through the protein kinase A signaling pathway in experiments with chicken chondrocytes in vitro [62, 63]. Our results indicate that PPP2CA levels are lowered in the medial secretome of the LM individual and not present in the HM secretomes. This could indicate that loss of PPP2CA is an early OA marker in the medial compartment of the LM individual that is completely lost in the end stage of the disease. However, the actual role of PPP2CA in OA needs to be further investigated.
Conclusions
We have demonstrated that proteins detected in the monolayer cultured chondrocyte secretome partly mirrors the proteins detected in the explant secretome and allows for quantitative studies. By applying the SILAC technology we demonstrated that the relative abundance of proteins significantly differed between chondrocytes isolated from the lateral and medial femoral condyle of OA patients (represented by Enolase 3, Stabilin 1, Transferrin, DMD SPRR2C and DPYSL3). Furthermore, in the LM scored chondrocyte secretome there were several proteins with a high SILAC ratio between the femur medial and lateral compartment e.g. MMP2 and TIMP1, that in combination with the Mankin scores may suggest that the LM medial femoral condyle secretome represents an early OA. Such unique data has to our knowledge never been published. Thus the differently expressed proteins could be potential markers of early OA degradation e.g. PPP2CA, IGFBP7, EFEMP1, FSTL1, CHI3L1, IGFBP4, SPARC and B2M. The high SILAC ratios in LM scored chondrocyte secretomes are lost in the HM scored chondrocyte secretomes, which indicates a shift in protein expression as the HM lateral femur compartment becomes more affected by the upgraded OA. Also, our data confirm experimental findings in mice regarding the role of DMD and CYTL1 in OA. There are also additional unique proteins for the LM scored patient that could represent healthy cartilage or early OA and could have potential for future functional studies e.g. new drug targets. For highlighted findings see list of proteins highlighted in the discussion.
List of proteins highlighted in the discussion
Gene name
Markers of early OA
PPP2CA
IGFBP7
EFEMP1
FSTL1
CHI3L1
IGFBP4
SPARC
B2M
Regulators of cartilage regeneration and differentiation
B2M
TF
CYTL1
IGFBP4
Regulators of extra cellular matrix homeostasis
STAB1
DMD
TIMP1
TIMP2
MMP2
CST3
PCOLCE
S100a8
Material and methods
Patient data and cartilage harvest
Human cartilage material used in this study was acquired from seven female total knee replacement cases suffering from severe OA and one female patient with macroscopically healthy knee cartilage. From two individuals, 73 and 82 years old undertaking total knee replacement, cartilage explants were obtained from the medial and lateral femur condyles. Chondrocytes for monolayer cultures were separately extracted from the medial and lateral femur condyle of the remaining five total knee replacement patients, 63, 67, 69 and two 72 years of age. The sixth monolayer culture was performed from macroscopically healthy cartilage biopsies taken from a patient 70 years of age undertaking leg amputation due to sarcoma with a macroscopically healthy knee cartilage. All samples were provided by the orthopedic surgeon department at Sahlgrenska University Hospital in Gothenburg with patient informed consent. Cartilage donations were approved by the ethical committee at the Sahlgrenska Academy at the University of Gothenburg, Gothenburg, Sweden.
Biopsy preparation for Mankin classification
Biopsies were isolated and a section of each biopsy was fixated in Histofix (Histolab products, Gothenburg, Sweden) for 24 hours, dehydrated with serial baths of increasing ethanol concentrations and embedded in paraffin. The paraffin embedded biopsies were cut into 5 μm sections onto microscope glass slides (Superfrost Plus, Menzel-Gläser, Germany), deparaffinized, Saffranin O stained and examined in a light microscope (Nikon, Japan). The biopsies were subsequently classified according to the modified Mankin score where the inspection of the tidemark has been excluded [26]. Mankin score is presented as mean out of three individual blinded scoring sessions performed by three experienced technicians. The OA specimens achieved higher Mankin scores and are in the following text accordingly denoted high Mankin (HM) patients while the unaffected cartilage specimen is denoted Low Mankin scored (LM).
Cartilage explant culture
Cartilage explants from two individuals were used for investigation of the explant secretome. Individual specimens were collected from the medial and lateral femur condyles respectively. Samples were cut into small pieces (approximately 2 mm3) and cultured for SILAC analysis. Femoral medial explants were incubated in SILAC DMEM/F12 medium containing 0.1 mg/mL of stable isotope labeled (heavy) arginine and lysine (U-13C6-Arginine and U-13C6-Lysine, Invitrogen) while femoral lateral explants were incubated in SILAC DMEM/F12 supplemented with 0.1 mg/mL normal (light) arginine and lysine (Invitrogen). The explant medium was changed daily for 10 days. Equal volumes (2 mL) of the conditioned explant culture medium from stable isotope labeled and unlabeled cultures were mixed prior to proteomic analysis. Duplicate samples of medium collected at day 2 and 10 from each patient were analyzed resulting in eight data-files representing the explant secretome.
Stable isotope labeling of chondrocytes in monolayer culture
Chondrocytes were separately extracted, as described earlier [6], from the medial and lateral femur condyle of five HM-scored patients affected with severe OA and from one LM-scored individual with macroscopically healthy knee cartilage. The extracted chondrocytes were seeded, at 16 × 103 cells/cm2, in SILAC DMEM/F12 medium (HM 1–5 and LM Experiment I). For the six chondrocyte cultures from the medial femur condyle the SILAC DMEM/F12 medium contained 0.1 mg/mL of stable isotope labeled arginine and lysine (U-13C6-Arginine and U-13C6-Lysine, Invitrogen) and for the six chondrocyte cultures from the lateral femur condyle the SILAC DMEM/F12 medium contained 0.1 mg/mL of normal arginine and lysine (Invitrogen). In addition, the LM scored cells were also cultured with the inverted labels (LM experiment II). Medium was changed every third day. All cultures were conducted in 37°C, 95% humidity and 5% CO2.
Protein enrichment and digestion
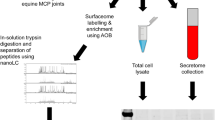
Supernatants from the cartilage explant cultures were collected after 2 and 10 days, respectively. Two aliquots of 2 mL from each time point were concentrated to 50 μL and the proteins were separated on SDS-PAGE gels as described below. After monolayer expansion of chondrocytes in SILAC-labeled (13C6-Lysine and 13C6-Arginine) or SILAC-unlabeled (12C6-Lysine and 12C6-Arginine) media for 5–6 cell doublings cells were conditioned in serum-free medium for 24 hours and subsequently the serum free conditioned media were collected and processed prior to MS-analysis. Equal volumes (2 mL) of the conditioned cell culture medium from SILAC-labeled and unlabeled cultures were mixed and the volume was reduced from 4 mL to approximately 50 μL on the Amicon Ultra-2 centrifugal concentrators (Millpore) with a molecular cut-off of 3,000 Da. The concentrated protein secretome in the retentate was separated by denaturing one-dimensional polyacrylamide gel electrophoresis (1-D SDS-PAGE). The concentrated protein solution (25 μL) was supplemented with NuPage LDS-sample buffer (Invitrogen) and dithriothreitol to a final concentration of 50 mM. Proteins were reduced and denatured by heating at 70°C for 10 minutes and loaded onto 4-12% Bis-Tris (NuPAGE® Novex) precast polyacrylamide gels, post-separation, proteins were visualized by Coomassie colloidal blue staining (Invitrogen). For in-gel trypsin digestion each gel lane was divided into 15 equally sized gel slices and subjected to automated trypsin digestion on the BioMek 2000 workstation equipped with a vacuum manifold. 96-well plates supplemented with a 7 μL volume of C18 reversed phase chromatographic resin (Lab-in-a-Plate Flow-Thru Plate, Glygen) were used for vacuum filtration and sample clean-up. The work-flow essentially followed the protocol previously described [64] except that the peptide extraction was performed twice with 0.2% trifluoroacetic acid (TFA) to allow for peptide binding to the C18 resin of the filter plates. Finally, peptides were eluted in two times 40 μL of 60% acetonitrile in 0.1% TFA and the eluted fractions were evaporated to dryness in a speedvac centrifuge. Prior to LC-MS/MS analysis samples were re-dissolved in 0.1% formic acid. For an experimental line-up see Figure 4.
Representative biopsy images and experimental line-up. Saffranin O staining of the LM lateral femur condyle (A), LM medial femur condyle (B), representative HM lateral femur condyle (D) and representative HM medial femoral condyle (E). Schematic view of the experimental procedure where chondrocytes were extracted from lateral and medial femoral condyle of the same knee joint, labeled in culture, incubated for medium conditioning and analyzed with MS (C).
LC-MS/MS analysis
Tryptic peptides, from digestion of the gel separated secretome, were separated on a 15 cm capillary column (Zorbax SB300 C18, 0.075 mm ID) by reversed phase chromatography. Peptides were reconstituted in 0.1% formic acid, loaded onto the column in eluent A (0.1% formic acid) and separated with a linear gradient from 3% to 60% eluent B (84% acetonitrile in 0.1% formic acid) at a flow rate of 250–300 nL/min. Gradient lengths were 130 min and the LC system (Ettan MDLC, GE Healthcare Life Sciences, NJ, USA) was directly coupled in-line with a LTQ-FTICR instrument (Thermo Fisher Scientific) via a nano-electrospray source (Thermo Fisher Scientific). The source was operated at 1.4 kV, with no sheath gas flow and with the ion transfer tube at 200°C. The mass spectrometer was programmed for acquisition in a data dependent mode. The survey scans were acquired in the FTICR mass analyzer at a resolution of 50,000 at m/z 400 and covered the m/z range 300–2,000, the 7 most intense peaks in each full mass scan, with charge state ≥ 2 and intensity above a threshold of 100, were selected for fragmentation in the linear ion trap (LTQ) by collision induced dissociation (CID). CID fragmentation was performed with normalized collision energy of 35%, activation q = 0.25, activation time of 30 ms and three microscans were averaged. For all fragmentation events dynamic exclusion was enabled with a repeat count of 2, peaks selected for fragmentation more than twice within 30 s were excluded from selection (20 p.p.m. window) for 180 seconds, the maximum number of excluded peaks was 200.
Protein identification and quantification
Raw data containing full-scan spectra acquired in profile mode and centroid MS/MS spectra, from the analysis of tryptic peptides, were processed using the MaxQuant software version 1.2.0.18 [65]. Default settings were used for feature extraction and first search for recalibration was performed against the human first search database provided with the software. The Andromeda search engine [66] integrated into the MaxQuant package was used for peptide identification and searches were performed against the IPI human database (version 3.68, 8,7061 sequences). Parameters for identification and quantification were set as follows: variable modification; oxidation of methionine and acetylation of the N-terminal, fixed modification; carbamidomethylation of cysteine, MS/MS tolerance 0.5 Da, peptide and protein FDR was set to 0.01 and for SILAC labeled samples the heavy label was set to arg6 and lys6. SILAC protein ratios are determined as the median of all peptide ratios assigned to the protein. For quantification a minimum peptide ratio count of two was set for each protein. To ensure that the Log2 values of the normalized protein H/L ratios followed a normal distribution and were centered around zero, histograms were plotted. Calculation of Significance B was done on the Log2 values of protein H/L ratios and intensities using the Perseus module (version 1.2.0.17) available in the MaxQuant environment. Threshold value for the significance B was set to 0.05 and the truncation was based on the Benjamini-Hochberg FDR.
Comparison of datasets
MaxQuant result files (ProteinGroups.txt and peptides.txt), including information on identified proteins and peptides were imported into the ProteinCenter software version 3.8.2014 (Proxeon Bioinformatics, Odense, Denmark). Data were filtered so that each identified protein contained at least 2 unique peptides and identified proteins were clustered, based on peptide sharing, into groups of indistinguishable proteins. All protein identifications originating from the analysis of culture media from explant incubations (eight samples) were merged into one single data set and all protein identifications from monolayer cultures (five samples) were merged into a second data set. Comparison of the two datasets was performed in the Protein Center software. An additional comparison was made to 100 proteins identified from monolayer-cultured chondrocytes in a study by Polacek et al. [22]. These protein identifiers were manually added to the ProteinCenter software.
Gene ontology analysis
The proteins that were differentially abundant when comparing the medial and lateral HM femoral condyle and the differentially abundant proteins when comparing the medial and lateral LM femoral condyle were put in a biological context by gene ontology (GO) analysis using Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 [67, 68]. The protein lists were uploaded to DAVID with official gene symbol as identifier and the gene ontology term Biological Process FAT (GOTERM_BP_FAT) was chosen for gene ontology analysis. Functional annotation chart was used for analyzing the functional annotation results. Genetic association database for disease was used in DAVID to analyze if the proteins common for both monolayer chondrocytes and explants were associated with any known disease.
Verification of MS quantifications by Enzyme Linked Immunosorbent Assay (ELISA)
TIMP metallopeptidase inhibitor 1 (TIMP1) and matrix metalloproteinase-2 (MMP2) protein levels in the chondrocyte-conditioned media used for SILAC analysis were determined with ELISA according to the manufactures recommendations (R&D Systems, MN, USA). The samples were diluted 1:49 for the TIMP1 ELISA and used undiluted for the MMP2 ELISA. Optical density of each well was determined with an Infinite® F50 Absorbance microplate reader set to measure 450 nm with the wavelength correction set to 540 nm (TECAN, Männedorf, Switzerland). Measurement data and protein amounts were calculated using the Magellan™ V6.6 software (TECAN).
References
Dillon CF, Rasch EK, Gu Q, Hirsch R: Prevalence of knee osteoarthritis in the United States: arthritis data from the Third National Health and Nutrition Examination Survey 1991–94. J Rheumatol 2006, 33: 2271–2279.
Muraki S, Oka H, Akune T, Mabuchi A, En-yo Y, Yoshida M, Saika A, Suzuki T, Yoshida H, Ishibashi H, Yamamoto S, Nakamura K, Kawaguchi H, Yoshimura N: Prevalence of radiographic knee osteoarthritis and its association with knee pain in the elderly of Japanese population-based cohorts: the ROAD study. Osteoarthr Cartil 2009, 17: 1137–1143. 10.1016/j.joca.2009.04.005
van Saase JL, van Romunde LK, Cats A, Vandenbroucke JP, Valkenburg HA: Epidemiology of osteoarthritis: Zoetermeer survey. Comparison of radiological osteoarthritis in a Dutch population with that in 10 other populations. Ann Rheum Dis 1989, 48: 271–280. 10.1136/ard.48.4.271
Felson DT, Naimark A, Anderson J, Kazis L, Castelli W, Meenan RF: The prevalence of knee osteoarthritis in the elderly. The Framingham Osteoarthritis Study. Arthritis Rheum 1987, 30: 914–918. 10.1002/art.1780300811
Neame R, Zhang W, Deighton C, Doherty M, Doherty S, Lanyon P, Wright G: Distribution of radiographic osteoarthritis between the right and left hands, hips, and knees. Arthritis Rheum 2004, 50: 1487–1494. 10.1002/art.20162
Brittberg M, Lindahl A, Nilsson A, Ohlsson C, Isaksson O, Peterson L: Treatment of deep cartilage defects in the knee with autologous chondrocyte transplantation. N Engl J Med 1994, 331: 889–895. 10.1056/NEJM199410063311401
Peterson L, Vasiliadis HS, Brittberg M, Lindahl A: Autologous chondrocyte implantation: a long-term follow-up. Am J Sports Med 2010, 38: 1117–1124. 10.1177/0363546509357915
Iliopoulos D, Malizos KN, Oikonomou P, Tsezou A: Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS One 2008, 3: e3740. 10.1371/journal.pone.0003740
Ruiz-Romero C, Calamia V, Mateos J, Carreira V, Martínez-Gomariz M, Fernández M, Blanco FJ: Mitochondrial dysregulation of osteoarthritic human articular chondrocytes analyzed by proteomics: a decrease in mitochondrial superoxide dismutase points to a redox imbalance. Mol Cell Proteomics 2009, 8: 172–189. 10.1074/mcp.M800292-MCP200
Ruiz-Romero C, Carreira V, Rego I, Remeseiro S, López-Armada MJ, Blanco FJ: Proteomic analysis of human osteoarthritic chondrocytes reveals protein changes in stress and glycolysis. Proteomics 2008, 8: 495–507. 10.1002/pmic.200700249
Wu J, Liu W, Bemis A, Wang E, Qiu Y, Morris EA, Flannery CR, Yang Z: Comparative proteomic characterization of articular cartilage tissue from normal donors and patients with osteoarthritis. Arthritis Rheum 2007, 56: 3675–3684. 10.1002/art.22876
Yammani RR, Long D, Loeser RF: Interleukin-7 stimulates secretion of S100A4 by activating the JAK/STAT signaling pathway in human articular chondrocytes. Arthritis Rheum 2009, 60: 792–800. 10.1002/art.24295
Huang J, Ballou LR, Hasty KA: Cyclic equibiaxial tensile strain induces both anabolic and catabolic responses in articular chondrocytes. Gene 2007, 404: 101–109. 10.1016/j.gene.2007.09.007
Sellam J, Berenbaum F: The role of synovitis in pathophysiology and clinical symptoms of osteoarthritis. Nat Rev Rheumatol 2010, 6: 625–635. 10.1038/nrrheum.2010.159
Ushiyama T, Chano T, Inoue K, Matsusue Y: Cytokine production in the infrapatellar fat pad: another source of cytokines in knee synovial fluids. Ann Rheum Dis 2003, 62: 108–112. 10.1136/ard.62.2.108
Abramson SB, Attur M: Developments in the scientific understanding of osteoarthritis. Arthritis Res Ther 2009, 11: 227. 10.1186/ar2655
Karlsson C, Brantsing C, Egell S, Lindahl A: Notch1, Jagged1, and HES5 are abundantly expressed in osteoarthritis. Cells Tissues Organs 2008, 188: 287–298. 10.1159/000121610
Bonin-Debs AL, Boche I, Gille H, Brinkmann U: Development of secreted proteins as biotherapeutic agents. Expert Opin Biol Ther 2004, 4: 551–558. 10.1517/14712598.4.4.551
Panoutsopoulou K, Southam L, Elliott KS, Wrayner N, Zhai G, Beazley C, Thorleifsson G, Arden NK, Carr A, Chapman K, Deloukas P, Doherty M, McCaskie A, Ollier WER, Ralston SH, Spector TD, Valdes AM, Wallis GA, Wilkinson JM, Arden E, Battley K, Blackburn H, Blanco FJ, Bumpstead S, Cupples LA, Day-Williams AG, Dixon K, Doherty SA, Esko T, Evangelou E, et al.: Insights into the genetic architecture of osteoarthritis from stage 1 of the arcOGEN study. Ann Rheum Dis 2011, 70: 864–867. 10.1136/ard.2010.141473
arcOGEN Consortium and arcOGEN Collaborators: Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet 2012, 380: 815–823.
Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M: Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 2002, 1: 376–386. 10.1074/mcp.M200025-MCP200
Polacek M, Bruun J-A, Johansen O, Martinez I: Differences in the secretome of cartilage explants and cultured chondrocytes unveiled by SILAC technology. J Orthop Res 2010, 28: 1040–1049.
Calamia V, Rocha B, Mateos J, Fernández-Puente P, Ruiz-Romero C, Blanco FJ: Metabolic Labeling of Chondrocytes for the Quantitative Analysis of the Interleukin-1-beta-mediated Modulation of Their Intracellular and Extracellular Proteomes. J Proteome Res 2011, 10: 3701–3711. 10.1021/pr200331k
Calamia V, Fernández-Puente P, Mateos JU, Lourido L, Rocha B, Montell E, Vergés J, Ruiz-Romero C, Blanco FJ: Pharmacoproteomic study of three different chondroitin sulfate compounds on intracellular and extracellular human chondrocyte proteomes. Mol Cell Proteomics 2011, 11: M111 013417.
Polacek M, Bruun J-A, Elvenes J, Figenschau Y, Martinez I: The secretory profiles of cultured human articular chondrocytes and mesenchymal stem cells: implications for autologous cell transplantation strategies. Cell Transplant 2011, 20: 1381–1393. 10.3727/096368910X550215
Mankin HJ, Dorfman H, Lippiello L, Zarins A: Biochemical and metabolic abnormalities in articular cartilage from osteo-arthritic human hips. II. Correlation of morphology with biochemical and metabolic data. J Bone Joint Surg Am 1971, 53: 523–537.
van der Sluijs JA, Geesink RGT, van der Linden AJ, Bulstra SK, Kuyer R, Drukker J: The reliability of the mankin score for osteoarthritis. J Orthop Res 1992, 10: 58–61. 10.1002/jor.1100100107
Kzhyshkowska J, Workman G, Cardó-Vila M, Arap W, Pasqualini R, Gratchev A, Krusell L, Goerdt S, Sage EH: Novel function of alternatively activated macrophages: stabilin-1-mediated clearance of SPARC. J Immunol 2006, 176: 5825–5832. (Baltimore, Md : 1950)
Heinegård D, Oldberg A: Structure and biology of cartilage and bone matrix noncollagenous macromolecules. FASEB J 1989, 3: 2042–2051.
Nakamura S, Kamihagi K, Satakeda H, Katayama M, Pan H, Okamoto H, Noshiro M, Takahashi K, Yoshihara Y, Shimmei M, Okada Y, Kato Y: Enhancement of SPARC (osteonectin) synthesis in arthritic cartilage. Increased levels in synovial fluids from patients with rheumatoid arthritis and regulation by growth factors and cytokines in chondrocyte cultures. Arthritis Rheum 1996, 39: 539–551. 10.1002/art.1780390402
Nanba Y, Nishida K, Yoshikawa T, Sato T, Inoue H, Kuboki Y: Expression of osteonectin in articular cartilage of osteoarthritic knees. Acta Med Okayama 1997, 51: 239–243.
Carlevaro MF, Albini A, Ribatti D, Gentili C, Benelli R, Cermelli S, Cancedda R, Cancedda FD: Transferrin promotes endothelial cell migration and invasion: implication in cartilage neovascularization. J Cell Biol 1997, 136: 1375–1384. 10.1083/jcb.136.6.1375
Vermeij WP, Florea BI, Isenia S, Alia A, Brouwer J, Backendorf C: Proteomic Identification of in Vivo Interactors Reveals Novel Function of Skin Cornification Proteins. J Proteome Res 2012, 11: 3068–3076. 10.1021/pr300310b
Tanaka H, Morimura R, Ohshima T: Dpysl2 (CRMP2) and Dpysl3 (CRMP4) phosphorylation by Cdk5 and DYRK2 is required for proper positioning of Rohon-Beard neurons and neural crest cells during neurulation in zebrafish. Dev Biol 2012, 370: 223–236. 10.1016/j.ydbio.2012.07.032
Nye DJ, Costas JM, Henley JB, Kim J-K, Plochocki JH: The chondrogenic response to exercise in the proximal femur of normal and mdx mice. BMC Musculoskelet Disord 2010, 11: 198. 10.1186/1471-2474-11-198
Barrett AJ, Davies ME, Grubb A: The place of human gamma-trace (cystatin C) amongst the cysteine proteinase inhibitors. Biochem Biophys Res Commun 1984, 120: 631–636. 10.1016/0006-291X(84)91302-0
Martel-Pelletier J, Cloutier JM, Pelletier JP: Cathepsin B and cysteine protease inhibitors in human osteoarthritis. J Orthop Res 1990, 8: 336–344. 10.1002/jor.1100080305
Mott JD, Thomas CL, Rosenbach MT, Takahara K, Greenspan DS, Banda MJ: Post-translational proteolytic processing of procollagen C-terminal proteinase enhancer releases a metalloproteinase inhibitor. J Biol Chem 2000, 275: 1384–1390. 10.1074/jbc.275.2.1384
Zhang H, Liew CC, Marshall KW: Microarray analysis reveals the involvement of beta-2 microglobulin (B2M) in human osteoarthritis. Osteoarthritis Cartilage 2002, 10: 950–960. 10.1053/joca.2002.0850
Stenhamre H, Slynarski K, Petrén C, Tallheden T, Lindahl A: Topographic variation in redifferentiation capacity of chondrocytes in the adult human knee joint. Osteoarthr Cartil 2008, 16: 1356–1362. 10.1016/j.joca.2008.03.025
Wakabayashi T, Matsumine A, Nakazora S, Hasegawa M, Iino T, Ota H, Sonoda H, Sudo A, Uchida A: Fibulin-3 negatively regulates chondrocyte differentiation. Biochem Biophys Res Commun 2010, 391: 1116–1121. 10.1016/j.bbrc.2009.12.034
Henrotin Y, Gharbi M, Mazzucchelli G, Dubuc J-E, De Pauw E, Deberg M: Fibulin 3 peptides Fib3–1 and Fib3–2 are potential biomarkers of osteoarthritis. Arthritis Rheum 2012, 64: 2260–2267. 10.1002/art.34392
Wang Y, Li D, Xu N, Tao W, Zhu R, Sun R, Fan W, Zhang P, Dong T, Yu L: Follistatin-like protein 1: a serum biochemical marker reflecting the severity of joint damage in patients with osteoarthritis. Arthritis Res Ther 2011, 13: R193. 10.1186/ar3522
Johansen JS, Jensen HS, Price PA: A new biochemical marker for joint injury. Analysis of YKL-40 in serum and synovial fluid. Br J Rheumatol 1993, 32: 949–955. 10.1093/rheumatology/32.11.949
Huang K, Wu LD: YKL-40: a potential biomarker for osteoarthritis. J Int Med Res 2009, 37: 18–24. 10.1177/147323000903700102
Kawahara C, Forster T, Chapman K, Carr A, Loughlin J: Genetic association analysis of the IGFBP7, ADAMTS3, and IL8 genes as the potential osteoarthritis susceptibility that maps to chromosome 4q. Ann Rheum Dis 2005, 64: 474–476.
Zhu W, Shiojima I, Ito Y, Li Z, Ikeda H, Yoshida M, Naito AT, Nishi J-I, Ueno H, Umezawa A, Minamino T, Nagai T, Kikuchi A, Asashima M, Komuro I: IGFBP-4 is an inhibitor of canonical Wnt signalling required for cardiogenesis. Nature 2008, 454: 345–349. 10.1038/nature07027
Blom AB, van Lent PL, van der Kraan PM, van den Berg WB: To seek shelter from the WNT in osteoarthritis? WNT-signaling as a target for osteoarthritis therapy. Current drug targets 2010, 11: 620–629. 10.2174/138945010791011901
Miclea RL, Siebelt M, Finos L, Goeman JJ, Löwik CWGM, Oostdijk W, Weinans H, Wit JM, Robanus-Maandag EC, Karperien M: Inhibition of Gsk3β in cartilage induces osteoarthritic features through activation of the canonical Wnt signaling pathway. Osteoarthr Cartil 2011, 19: 1363–1372. 10.1016/j.joca.2011.07.014
Corr M: Wnt-beta-catenin signaling in the pathogenesis of osteoarthritis. Nat Clin Pract Rheumatol 2008, 4: 550–556.
Kim J-S, Ryoo ZY, Chun J-S: Cytokine-like 1 (Cytl1) regulates the chondrogenesis of mesenchymal cells. J Biol Chem 2007, 282: 29359–29367. 10.1074/jbc.M700965200
Jeon J, Oh H, Lee G, Ryu J-H, Rhee J, Kim J-H, Chung K-H, Song W-K, Chun C-H, Chun J-S: Cytokine-like 1 knock-out mice (Cytl1-/-) show normal cartilage and bone development but exhibit augmented osteoarthritic cartilage destruction. J Biol Chem 2011, 286: 27206–27213. 10.1074/jbc.M111.218065
Zreiqat H, Belluoccio D, Smith MM, Wilson R, Rowley LA, Jones K, Ramaswamy Y, Vogl T, Roth J, Bateman JF, Little CB: S100A8 and S100A9 in experimental osteoarthritis. Arthritis Res Ther 2010, 12: R16. 10.1186/ar2917
Lee S-W, Jung KH, Jeong CH, Seo JH, Yoon D-K, Suh J-K, Kim K-W, Kim WJ: Inhibition of endothelial cell migration through the down-regulation of MMP-9 by A-kinase anchoring protein 12. Mol Med Rep 2011, 4: 145–149.
Suehara Y, Tochigi N, Kubota D, Kikuta K, Nakayama R, Seki K, Yoshida A, Ichikawa H, Hasegawa T, Kaneko K, Chuman H, Beppu Y, Kawai A, Kondo T: Secernin-1 as a novel prognostic biomarker candidate of synovial sarcoma revealed by proteomics. J Proteomics 2011, 74: 829–842. 10.1016/j.jprot.2011.02.033
Way G, Morrice N, Smythe C, O’Sullivan AJ: Purification and identification of secernin, a novel cytosolic protein that regulates exocytosis in mast cells. Mol Biol Cell 2002, 13: 3344–3354. 10.1091/mbc.E01-10-0094
Miyoshi N, Ishii H, Mimori K, Sekimoto M, Doki Y, Mori M: SCRN1 is a novel marker for prognosis in colorectal cancer. J Surg Oncol 2010, 101: 156–159.
Zhu C, Hu DL, Liu YQ, Zhang QJ, Chen FK, Kong XQ, Cao KJ, Zhang JS, Qian LM: Fabp3 inhibits proliferation and promotes apoptosis of embryonic myocardial cells. Cell Biochem Biophys 2011, 60: 259–266. 10.1007/s12013-010-9148-2
Song G-X, Shen Y-H, Liu Y-Q, Sun W, Miao LP, Zhou L-J, Liu H-L, Yang R, Kong X-Q, Cao KJ, Qian L-M, Sheng Y-H: Overexpression of FABP3 promotes apoptosis through inducing mitochondrial impairment in embryonic cancer cells. J Cell Biochem 2012, 113: 3701–3708. 10.1002/jcb.24243
Shen Y-H, Song G-X, Liu Y-Q, Sun W, Zhou L-J, Liu H-L, Yang R, Sheng Y-H, Qian L-M, Kong X-Q: Silencing of FABP3 promotes apoptosis and induces mitochondrion impairment in embryonic carcinoma cells. J Bioenerg Biomembr 2012, 44: 317–323. 10.1007/s10863-012-9439-y
Lires-Deán M, Caramés B, Cillero-Pastor B, Galdo F, López-Armada MJ, Blanco FJ: Anti-apoptotic effect of transforming growth factor-beta1 on human articular chondrocytes: role of protein phosphatase 2A. Osteoarthr Cartil 2008, 16: 1370–1378. 10.1016/j.joca.2008.04.001
Zákány R, Bakó E, Felszeghy S, Holló K, Balázs M, Bárdos H, Gergely P, Módis L: Okadaic acid-induced inhibition of protein phosphatase 2A enhances chondrogenesis in chicken limb bud micromass cell cultures. Anat Embryol (Berl) 2001, 203: 23–34. 10.1007/s004290000128
Zákány R, Szucs K, Bakó E, Felszeghy S, Czifra G, Bíró T, Módis L, Gergely P: Protein phosphatase 2A is involved in the regulation of protein kinase A signaling pathway during in vitro chondrogenesis. Exp Cell Res 2002, 275: 1–8. 10.1006/excr.2002.5487
Forsman A, Rüetschi U, Ekholm J, Rymo L: Identification of Intracellular Proteins Associated with the EBV-Encoded Nuclear Antigen 5 Using an Efficient TAP Procedure and FT-ICR Mass Spectrometry. J Proteome Res 2008, 7: 2309–2319. 10.1021/pr700769e
Cox J, Mann M: MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008, 26: 1367–1372. 10.1038/nbt.1511
Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M: Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 2011, 10: 1794–1805. 10.1021/pr101065j
Huang DW, Sherman BT, Lempicki RA: Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009, 37: 1–13. 10.1093/nar/gkn923
Huang DW, Sherman BT, Lempicki RA: Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009, 4: 44–57.
Acknowledgements
We thank Associate Professor Sven Inerot at the department of Orthopaedics at the Sahlgrenska University Hospital for providing cartilage biopsies and his input on the study design. We thank Ann-Charlotte Westerlund and all surgeons at the department of Orthopeadics for providing, and the excellent handling of, the biopsy material. We also thank Erna Koskel with personnel for the excellent handling of the patient informed consent.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JS and UR designed the study, carried out the cell cultures, performed and analyzed the proteomic analysis together with the statistic analysis and drafted the manuscript. ES and JK participated in the study design and helped to draft the manuscript. AL conceived the study, analyzed the proteomic data and participated in drafting the manuscript. All authors have read and approved the final manuscript.
Johan Stenberg, Ulla Rüetschi contributed equally to this work.
Electronic supplementary material
12953_2013_452_MOESM1_ESM.xlsx
Additional file 1: Table S1: Proteins identified in cellmedium from explant cultures of two OA patient samples (Patient 1 and Patient 2). Cellmedia collected at day 2 and 10 were collected and analysed in duplicates. (XLSX 104 KB)
12953_2013_452_MOESM2_ESM.xlsx
Additional file 2: Table S2: Proteins identified in cellmedium from monolayer cultures of five OA patient samples. (XLSX 252 KB)
12953_2013_452_MOESM3_ESM.xlsx
Additional file 3: Table S3: Proteins common for the combined OA explant seretome and the combined high Mankin scored monolayer secretomes. (XLSX 16 KB)
12953_2013_452_MOESM5_ESM.xlsx
Additional file 5: Table S5: Proteins identified in cellmedium from monolayer cultured chondrocytes of one LM-scored patient sample. Duplicate samples were individually SILAC labelled and processed in parallel. LM Exp I = Low Mankin label experiment one, LM Exp II = Low Mankin label experiment two. (XLSX 132 KB)
12953_2013_452_MOESM6_ESM.xlsx
Additional file 6: Table S6: All medially/latterally regulated proteins in the HM secretomes. Silac protein ratio>1= Femur medial abundance. Silac protein ratio<1= Femur lateral abundance. HM=High Mankin scored individual. NR= No Ratio. (XLSX 22 KB)
12953_2013_452_MOESM7_ESM.xlsx
Additional file 7: Table S7: Up-regulated proteins in the LM femur lateral secretome. LM I=Low Mankin scored individual experiment one. LM II=Low Mankin scored individual experiment two. HM.1-5=High Mankin scored individual 1-5. NR=No Ratio. (XLSX 36 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Stenberg, J., Rüetschi, U., Skiöldebrand, E. et al. Quantitative proteomics reveals regulatory differences in the chondrocyte secretome from human medial and lateral femoral condyles in osteoarthritic patients. Proteome Sci 11, 43 (2013). https://doi.org/10.1186/1477-5956-11-43
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1477-5956-11-43