Abstract
Background
An important inter-individual variability in the response of insulin sensitivity following a fish oil supplementation has been observed. The objective was to examine the associations between single nucleotide polymorphisms (SNPs) within sterol regulatory element binding transcription factor 1 (SREBF1) gene and the response of insulin sensitivity to a fish oil supplementation.
Methods
Participants (n = 210) were recruited in the greater Quebec City area and followed a 6-week fish oil supplementation protocol (5 g/day: 1.9-2.2 g EPA; 1.1 g DHA). Insulin sensitivity was assessed by the quantitative insulin sensitivity check index (QUICKI). Three tag SNPs (tSNPs) within SREBF1 gene were genotyped according to TAQMAN methodology.
Results
Three tSNPs (rs12953299, rs4925118 and rs4925115) covered 100% of the known genetic variability within SREBF1 gene. None of the three tSNPs was associated with either baseline fasting insulin concentrations (rs12953299, rs4925118 and rs4925115) (p = 0.29, p = 0.20 and p = 0.70, respectively) or QUICKI (p = 0.20, p = 0.18 and p = 0.76, respectively). The three tSNPs (rs12953299, rs4925118 and rs4925115) were associated with differences in the response of plasma insulin levels (p = 0.01, p = 0.005 and p = 0.004, respectively) and rs12953299 as well as rs4925115 were associated with the insulin sensitivity response (p = 0.009 and p = 0.01, respectively) to the fish oil supplementation, independently of the effects of age, sex and BMI.
Conclusions
The genetic variability within SREBF1 gene has an impact on the insulin sensitivity in response to a fish oil supplementation.
Trial registration
clinicaltrials.gov: NCT01343342.
Similar content being viewed by others
Introduction
The sterol regulatory element binding transcription factor 1 (SREBF1) gene encodes a transcription factor which is a main regulator of lipid metabolism, the sterol regulatory element-binding protein-1c (SREBP-1c)[1]. SREBF1 gene is expressed in multiple tissues including liver, white and brown adipose tissue, adrenal gland and to a lower extent in pancreatic β-cell[2, 3]. Insulin induces the expression of the SREBF1 gene in adipose tissue, liver and muscle cells[1]. However, in pancreatic β-cell, it has been observed that SREBP-1c modulates insulin secretion potentially through a mechanism involving lipotoxicity[3, 4]. SREBP-1c may be involved in reticulum endoplasmic stress and in β-cell apoptosis[5]. The knockdown of SREBP-1c in pancreatic β-cell inhibited the expression of markers of reticulum endoplasmic stress[5].
SREBF1 gene expression is also regulated by dietary intakes. For example, an insulin independent effect has been demonstrated with different types of sugar such as glucose, fructose and sucrose, on SREBF1 gene expression induction[6]. Dietary fats also affect SREBF1 gene expression; a high saturated fat (SFA) diet increases SREBF1 gene expression both in the liver and in pancreatic β-cell[3, 7] whereas a diet high in polyunsaturated fat (PUFA) decreases SREBF1 gene expression[3, 8, 9]. The intake of fish oil may have a favorable impact on insulin sensitivity. Among fructose-induced hypertriglyceridemic and insulin resistant male rhesus macaques, the intake of 4 g/day of fish oil prevented the development of hypertriglyceridemia and insulin resistance[10]. Studies observing rodents have also observed a beneficial effect of fish oil on insulin sensitivity[11, 12]. In diet induced obese mice, the intake of fish oil reduces SREBF1 gene expression levels in the liver and modifies the expression of other genes involved in lipid metabolism such as fatty acid synthase gene (FASN) and acyl-Coenzyme A oxidase 1 (ACOX1)[13]. Eicosapentaenoic acid (EPA) was shown to inhibit SREBP-1 maturation[14] and to restore insulin secretion after suppression by palmitate through an SREBP-1c dependent mechanism[15].
The genetic variability within the SREBF1 gene may play a role in insulin resistance or type 2 diabetes. A meta-analysis of genome wide scans in European populations showed linkage with type 2 diabetes in the 17p11 region, which comprises the SREBF1 gene[16]. Single nucleotide polymorphisms (SNPs) within the SREBF1 gene have been associated with type 2 diabetes, insulin resistance, obesity and blood lipid levels[17–22]. Among humans the impacts of fish oil on insulin sensitivity, glucose concentrations and/or the risk of type 2 diabetes have not been consistent[23–27]. Some studies even observed an increase in fasting insulin concentrations and/or fasting glucose concentrations[28, 29]. These inconsistencies in results could be partly due to differences in the genetic background, dietary intakes and/or lifestyle. Our group previously observed a large inter-individual variability in the response of insulin sensitivity to a fish oil supplementation[30]. Thus, the objective of this study was to examine the associations between SNPs within SREBF1 gene and the plasma insulin and glucose response to a fish oil supplementation.
Methods
Participants
Methods related to this study cohort have been previously described[31]. Briefly, a total of 254 unrelated participants from the greater Quebec City metropolitan area were recruited to participate in this clinical trial between September 2009 and December 2011 through advertisements in local newspapers as well as by electronic messages sent to university students/employees. To be eligible, participants had to be non-smokers and without any thyroid or metabolic disorders requiring treatment, for example diabetes, hypertension, severe dyslipidemia, and/or coronary heart disease. A total of 210 participants completed the fish oil supplementation period. However, fasting insulin and glucose concentrations were obtained only for 207 participants. The experimental protocol was approved by the ethics committees of Laval University Hospital Research Center and Laval University. This clinical trial was registered at clinicaltrials.gov (NCT01343342). Informed written consent was obtained from all the study participants.
Study design and diets
The study design and diets have been described previously[31]. Briefly, participants followed a run-in period of 2 weeks. Individual dietary instructions were given by a trained dietitian to achieve the recommendations from Canada’s Food Guide. After the 2-week run-in period, each participant received a bottle containing fish oil capsules for the next 6 weeks. They were instructed to take five capsules (1 g of fish oil/capsule) per day (Ocean Nutrition, Nova Scotia, Canada), providing a total of 5 g of fish oil (1.9-2.2 g EPA and 1.1 g docosahexaenoic acid (DHA)) per day. Compliance was assessed from the return of bottles and by measuring erythrocyte membranes and plasma phospholipids fatty acid (FA) composition. Dietary intakes were assessed at screening using a validated food frequency questionnaire (FFQ)[32]. Dietary intakes were also measured pre- and post-supplementation using two 3-day dietary records.
Biochemical parameters
The morning after a 12-hour overnight fast and 48-h alcohol abstinence, blood samples were collected from an antecubital vein into vacutainer tubes containing EDTA. Blood samples were collected at screening, baseline, pre-supplementation (two weeks after baseline) and post-supplementation (six weeks after pre-supplementation). Plasma was separated by centrifugation (2500 × g for 10 minutes at 4°C), samples were aliquoted and frozen for subsequent analyses. Plasma total cholesterol (total-C) and plasma triglyceride concentrations were measured using enzymatic assays[33, 34]. Infranatant (d >1.006 g/ml) with heparin-manganese chloride was used to precipitate VLDL and LDL and then determine HDL-cholesterol concentrations (HDL-C)[35]. The equation of Friedewald was used to estimate LDL-cholesterol concentrations (LDL-C)[36]. Non-HDL-C was calculated by subtracting HDL-C from total-C. Fasting insulin concentrations were measured by radioimmunoassay with polyethylene glycol separation[37]. Fasting glucose concentrations were enzymatically measured[38]. The quantitative insulin sensitivity check index (QUICKI) was used as a marker of insulin sensitivity calculated as follow: 1/(log(insulin(mU/L)) + log(glucose (mg/dL)))[39]. QUICKI has been reported to be more reproducible than the common homeostasis model assessment (HOMA) of insulin resistance[40] and has a strong linear correlation with glucose clamp estimates among various types of health conditions (healthy, obesity, insulin resistance, diabetes and hypertension)[41].
SNPs selection and genotyping
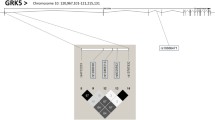
As described previously[31], SNPs were selected with the International HapMap Project SNP database (HapMap Data Rel 28 Phase II + III, August 10, on National Center for Biotechnology Information (NCBI) B36 assembly, dbSNP b126). Tag SNPs (tSNPs) were determined with the tagger procedure in HaploView software version 4.2 with minor allele frequency (MAF) of >0.05 and pairwise tagging r2 ≥ 0.80. Afterwards, as shown in Figure 1, linkage disequilibrium (LD) plot were generated with Haploview software version 4.2. Figure 1 also illustrates the high LD between the chosen tSNP rs4925115 and the well described rs2297508 located in exon 18c of SREBF1 gene[17, 20, 21]. All tSNPs were genotyped within INAF laboratories with the TAQMAN methodology[42], as described previously[43]. Briefly, genotypes were determined using ABI Prism SDS version 2.0.5 (Applied Biosystem, Foster City, CA, USA). All SNPs were successfully genotyped.
Linkage disequilibrium (LD) plot of tSNPs within SREBF1 gene. Figure legend. LD plots were generated by HaploView software version 4.2 using r2 LD values. Two of the selected tSNPs (rs12953299 and rs4925115 of the first cohort) were in moderate to high LD with the exonic SNP rs2297508 (second cohort) (rs12953299 (r2 = 0.62) and rs4925115 (r2 = 0.95)).
Fatty acid composition of erythrocyte membranes and plasma phospholipids
As described previously[44], FA composition was measured in erythrocyte membranes by gas chromatographic analysis in a subset of 31 participants. Methods to extract plasma phospholipids have been described elsewhere[31]. FA composition from plasma phospholipids were measured on the total cohort of 210 participants. Briefly, plasma lipids were extracted according to a modified Folch method[45]. Capillary gas chromatography was used to obtain FA profiles[46]. FA profiles both in erythrocyte membranes and plasma phospholipids were expressed as the relative percentage areas of total FAs.
Gene expression assessment
Blood samples (pre- and post- supplementation) were collected into an 8-ml Cell Preparation Tube (CPT) (Becton Dickinson, Oakville, On, Canada). Gene expression levels were measured in peripheral blood mononuclear cells (PBMCs), which are considered a valid proxy measure for many tissues including the liver[47, 48]. Peripheral blood mononuclear cells (PBMCs) were separated by centrifugation (1500 × g, 20 min, at room temperature) and washed according to the manufacturer’s instructions. Total RNA was extracted with RNeasy Plus Mini Kit (QIAGEN, Mississauga, On, Canada) according to manufacturer’s protocol. Spectrophotometric quantification was realised with NanoDrop 2000C UV–vis Spectrophotometer (Thermo Scientific) and cDNA was generated using 400 ng of total RNA with the High Capacity cDNA Reverse Transcription Kit (Life Technologies™). cDNA was mixed with TaqMan OpenArray® Real-Time PCR Master Mix (#4462164, Life Technologies™). The assays used were as follows: Hs01088691_m1 (SREBF1) and GAPDH Hs99999905_m1 as the housekeeping gene. All assays used the same fluorescent reporter probe (FAM dye labeled). All samples were run in triplicate on a QuantStudio™ 12 K Flex Real-Time PCR (RT-PCR) System (Life Technologies™) using 48-well plates TaqMan® OpenArray® RT PCR Inventoried Format 18. The RT-PCR results were analysed with ExpressionSuite software v1.0.1 (Life Technologies™).
Second cohort
Seven hundred (700) Caucasians aged between 18 and 55 years were recruited in the Quebec City metropolitan region. Recruitment occurred between 2004 and 2006 through public advertisements (local newspapers and electronic messages) sent to university and hospital employees, as described previously[49]. A trained research assistant took anthropometric measures. A registered dietitian administered a validated FFQ to assess dietary intakes[32]. SREBF1 c.*619C > G (rs2297508) was genotyped using the TAQMAN methodology[42]. Statistical analyses were performed by using a model including the interaction term SNP*PUFA with the GLM procedure in SAS and the type 3 sum of squares for unbalanced study design. Age, sex, BMI and total energy intakes were considered as confounding variables.
Statistical analyses
The Hardy-Weinberg equilibrium was tested with the ALLELE procedure of SAS version 9.3 using Fisher’s exact test (P < 0.01). When the genotype frequency for homozygous individuals of the minor allele was <5%, carriers (heterozygotes and homozygotes) of the minor allele were grouped.
The sample size was calculated based on plasma triglyceride changes following the fish oil supplementation with a genetic variation occurring in a relatively low frequency (5%) of the population. A group of 152 participants was sufficient to provide an 80% probability and a 5% significance level of detecting an anticipated difference of 0.25 mmol/L in plasma triglyceride concentrations after 6 weeks of fish oil supplementation.
Non-normally distributed variables were logarithmically transformed. Fasting insulin concentration values higher or lower than means ± 3 multiplied by standard deviation (SD) were considered as outliers (n = 6), thus 201 participants were kept for the statistical analyses. Differences were assessed using analyses of variance (ANOVA) with the GLM procedure in SAS and the type 3 sum of squares for unbalanced study design. The fasting insulin response (delta) was calculated as followed: ((post-supplementation insulin concentrations minus pre-supplementation insulin concentrations)/pre-supplementation insulin concentrations*100). The same model was used to test the associations with fasting glucose concentrations, insulin sensitivity (QUICKI) and FA composition both in erythrocytes and plasma phospholipid membranes. Each model was adjusted for the effects of age, sex and BMI. To take into account the impact of multiple testing, the simpleM method described by Gao et al.[50] was utilised. Briefly, this method considers the impacts of LD between SNPs and has been demonstrated as efficient and accurate comparatively to permutation-based corrections[50]. First, the composite LD correlation matrix was derived from the data set. Then, eigenvalues were calculated using the SAS PRINCOMP procedure and the number of effective independent tests was inferred so that the corresponding eigenvalues explain 99.5% of the variation in SNP data or the variables (fasting glucose, insulin and QUICKI), as proposed by Gao et al.[50]. According to Gao’s method, the number of effective independent tests for the three SNPs was 2 and for the three traits (fasting glucose, insulin and QUICKI) was 2. The final step applies the Bonferroni correction formula to calculate the adjusted point-wise significance level, which was defined as αG = 0.05/(2X2) (effective independent tests). Thus, p-values <0.0125 were considered significant (p = 0.05/(4)). All statistical analyses were performed using SAS statistical software version 9.3 (SAS Institute, Inc., Cary, NC, USA).
Results
All tSNPs were in Hardy-Weinberg equilibrium. Three tSNPs covered 100% of the known genetic variability within the SREBF1 gene[31]. As presented in Figure 1, two of the selected tSNPs were in moderate to high LD with the exonic SNP rs2297508 (rs12953299 (r2 = 0.62) and rs4925115 (r2 = 0.95)).
Baseline characteristics of the study participants are shown in Table 1. Before the fish oil supplementation period, no differences in fasting insulin concentrations according to genotypes were observed for the three tSNPs (rs12953299, rs4925118 and rs4925115) (p = 0.29, p = 0.20 and p = 0.70, respectively). Also there were no differences in either fasting glucose concentrations according to genotypes of rs12953299, rs4925118 and rs4925115 (p = 0.16, p = 0.64 and p = 0.22, respectively) or for insulin sensitivity (QUICKI) values (p = 0.20, p = 0.18 and p = 0.76, respectively).
Globally, during the study protocol fasting insulin concentrations were not modified (pre-supplementation insulin concentrations: 77.7 ± 29.3pmol/L; post-supplementation insulin concentrations: 79.0 ± 30.0pmol/L) (p = 0.52)). Fasting glucose concentrations slightly increased from 4.95 ± 0.44 mmol/L to 5.04 ± 0.49 mmol/L after the fish oil supplementation period (p = 0.0002), as previously reported[51]. An important inter-individual variability has been observed in the response of fasting insulin concentrations, ranging from a decrease of -53.0% to an increase of +135.2%. Briefly, 110 individuals decreased (relative change ≤ 0%) and 91 increased (relative change > 0%) their fasting insulin concentrations. Globally, the mean change in insulin concentrations was 5.1% ± 30.0%. The insulin sensitivity (QUICKI) was not modified by the fish oil supplementation (p = 0.19).
No differences according to genotypes of the SREBF1 gene were observed in the response of fatty acid n-3 PUFA (EPA, DHA and total n-3 PUFA) phospholipid content to the fish oil supplementation. A difference was observed for EPA concentrations in erythrocyte membranes according to rs4925118 genotypes (p = 0.02) for which the relative increase following the fish oil supplementation was greater among C/C homozygotes than for the T allele carriers (T/T + C/T = 147.7 ± 80.0% (n = 4); C/C = 236.5 ± 73.0% (n = 23)). A trend (p = 0.07) was also observed for pre-supplementation EPA content in erythrocyte membranes according to rs4925118 genotypes (T/T + C/T = 0.88 ± 0.33% (n = 4); C/C = 0.68 ± 0.22% (n = 24)).
SREBF1 gene expression levels were not modified by the fish oil supplementation (p = 0.85). As shown in Table 2, no differences were observed in the response of SREBF1 gene expression levels to fish oil supplementation between genotypes of rs4925118 and rs4925115 (p = 0.59, p = 0.47 and p = 0.25, respectively).
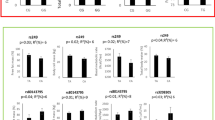
As shown in Figure 2, the% change of fasting insulin concentrations ((post-supplementation insulin concentrations minus pre-supplementation insulin concentrations)/pre-supplementation insulin concentrations*100) following the fish oil supplementation was different according to genotypes of the three tSNPs (rs12953299, rs4925118 and rs4925115) within the SREBF1 gene, adjusted for age, sex and BMI (p = 0.01, p = 0.005 and p = 0.004, respectively). The response of fasting glucose concentrations was not different according to genotypes of the tSNPs (rs12953299, rs4925118 and rs4925115). The insulin sensitivity (QUICKI) was associated with the tSNPs rs12953299 and rs4925115 (p = 0.009 and p = 0.01, respectively) but not with rs4925118 (p = 0.16). The potential effects of total dietary n-3 PUFA intakes in the participants’ habitual diets were included as a confounding variable in these models and did not modify the associations observed (data not shown).
The relative response in fasting insulin concentrations and QUICKI index (insulin sensitivity) according to genotype. Figure legend. a) rs12953299 (A/A: n = 46, A/G: n = 100, G/G: n = 55); Delta insulin (A/A: 15.3 ± 32.0%, A/G: 1.2 ± 30.1%, G/G: 3.9 ± 26.4%), P-value for delta insulin model: p = 0.01; Delta QUICKI (A/A: -2.0 ± 4.1%, A/G: 0.4 ± 4.8%, G/G: 3.9 ± 26.4%), P-value for delta QUICKI model: p = 0.009 b) rs4925118 (T/T + C/T: n = 67, C/C: n = 134); Delta insulin (T/T + C/T: 14.1 ± 36.2%, C/C: 0.6 ± 25.3%), P-value for delta insulin model: p = 0.005; P-value for delta QUICKI model: p = 0.16 c) rs4925115 (A/A: n = 33, A/G: n = 105, G/G: n = 63); Delta insulin (A/A: 19.5 ± 34.0%, A/G: 3.4 ± 29.3%, G/G: 0.6 ± 27.0%), P-value for delta insulin model: p = 0.004; Delta QUICKI (A/A: -2.3 ± 4.2%, A/G: -0.1 ± 4.5%, 0.4 ± 4.5%), P-value for delta QUICKI model: p = 0.01. Delta values (relative change) were calculated as ((post-supplementation values minus pre-supplementation values)/pre-supplementation values*100). All differences were assessed with ANOVA adjusted for age, sex and BMI. Means ± SE.
Second cohort
As presented in Table 3, one significant gene-diet interaction effects on QUICKI (insulin sensitivity index) was observed between rs2297508 and dietary PUFA intakes (in grams) (p = 0.05). To further understand these associations dietary PUFA intakes were divided in tertiles. Figure 3 presents QUICKI values according to genotype of rs2297508 and tertiles of dietary PUFA intakes. A significant difference was observed only among C/C homozygotes for which individuals with the highest dietary PUFA intakes had higher QUICKI values than individuals with the lowest dietary PUFA intakes (p = 0.03). A trend (p = 0.06) was observed for the interaction effect on QUICKI between rs2297508 and dietary intakes of omega-3 PUFA (in grams). Both models were adjusted for the effects of age, sex, BMI and energy intakes. Genotype of rs2297508 alone was not associated with QUICKI values (p = 0.21). No gene-diet interaction effects were observed on fasting insulin or glucose concentrations.
QUICKI index values according to rs2297508 genotype and tertiles of dietary PUFA intakes. Figure legend. Tertile 1 of dietary PUFA intakes (3.17 g-11.97 g) (G/G: n = 48; C/G: n = 95; C/C: n = 67), Tertile 2 of dietary PUFA intakes (11.98 g-16.49 g) (G/G: n = 41; C/G: n = 91; C/C: n = 82) and Tertile 3 of dietary PUFA intakes (16.53 g-48.18 g) (G/G: n = 27; C/G: n = 96; C/C: n = 86). Differences in QUICKI values between tertiles were assessed with an ANOVA by genotype adjusted for the effects of age, sex and BMI. Means with different letters are significantly different. Means ± SE.
Discussion
Participants of this cohort were overweight but generally considered as healthy according to lipid concentration values[52]. Fasting glucose concentrations were within normal values[53]. The QUICKI index indicated a probable borderline insulin resistance state among these participants[39, 41, 54]. An important inter-individual variability in the response of fasting insulin concentrations was observed. This wide inter-individual variability in HOMA-insulin sensitivity (IS) response in this cohort has been previously described[30]. In this study, effects of tSNPs within the SREBF1 gene on the fasting insulin and insulin sensitivity responses were observed after the fish oil supplementation.
SREBF1 gene is an important transcription factor regulating many genes involved in lipid metabolism and also in insulin induced glucose metabolism[1]. Moreover, the expression of SREBF1 gene is significantly affected by dietary intakes, including fish oil[3, 6–9, 13]. Therefore, SREBF1 gene is an interesting candidate for the study of inter-individual variability in the response of fasting insulin concentrations to a fish oil supplementation. Whether these impacts are mediated through the effects of SREBP-1c within hepatocytes and/or pancreatic cells is unknown. In the liver, increased SREBP-1c concentrations have been shown to repress the transcription of insulin receptor substrate 2 (IRS2) gene which led to a detrimental impact on insulin sensitivity, a fatty liver and a production of VLDL enriched in triglycerides[55]. IRS2 mediates insulin signaling in the liver[55]. Insulin in the liver activates glycogen synthesis, inhibits hepatic glucose output and promotes lipogenesis[55]. The induction of SREBP-1c in mice resulted in impaired secretion and glucose intolerance, as reviewed by Shimano et al.[3]. SREBP-1c may also affect insulin secretion of pancreatic β-cell through a mechanism involving uncoupling protein-2 (UCP2)[56]. A sterol regulatory element (SRE) has been discovered in the promoter region of UCP2 gene[56]. An increase in UCP2 gene expression is associated with a lower efficacy of glucose-induced insulin secretion[57]. In the present study no differences in SREBF1 gene expression were observed following the fish oil supplementation. Studies examining the impacts of PUFA, fish oil or EPA on SREBF1 gene regulation have been conducted among mice or in vitro with human cells[6–9, 13–15]. It is possible that the dose used for the supplementation in this study was insufficient to observe an effect on SREBF1 gene expression. However, the activity of lipogenic target genes of SREBP-1c transcription factor is not only regulated by SREBF1 mRNA abundance. For example, Tanaka et al.[14] did not observe reduced SREBF1 mRNA levels but rather an inhibition of the maturation of SREBP-1c. Thus, the fish oil supplementation in the present study may have had effects on the insulin response through posttranslational modifications of SREBP-1c and its impact on subsequent target genes. However, we cannot rule out the possibility that significant differences may have been observed if expression levels were directly measured in hepatocytes[58].
The overall effects of fish oil intakes on insulin resistance, glycemic control and the risk of type 2 diabetes appears to be negligible, as recently reviewed by Wu et al.[59]. However, the authors observed a large heterogeneity. Thus, it is possible that for some individuals the impacts of fish oil intake on the risk of type 2 diabetes or other related biologic parameters may be beneficial and for some other individuals detrimental. Quite a few studies have observed a modest increase in fasting glucose concentrations after fish oil intake[60]. It has been observed that the reduction in plasma triglyceride concentrations induced by n-3 PUFA intake may be partly induced by the increased use of glycerol for gluconeogenesis which may explain increases in fasting glucose concentrations[61, 62]. The increase in fasting glucose concentrations was also observed in this cohort[51]. However, because this study was designed without a control group, we cannot rule out the possibility that part of these changes may be related to modifications in the participants’ lifestyle during the protocol. Still, participants were asked to maintain their nutritional and physical activity habits stable during the intervention. Assessment of dietary intakes and physical activity levels during the study protocol revealed that they were quite stable (data not shown).
In the present study, homozygotes for minor alleles of the tSNPs rs12953299 and rs4925115 were associated with an increase in fasting insulin concentrations and a decrease in insulin sensitivity assessed by QUICKI after a fish oil supplementation compared to the other genotypes. For the tSNP rs4925118 only a difference in the fasting insulin response was observed, carriers of the T allele increased their fasting insulin concentrations after a fish oil supplementation compared to C/C homozygotes. Thus, for these genotypes the impact of fish oil on insulin sensitivity may be detrimental. For the other genotypes, the fish oil supplementation had a minor impact which may be less likely to increase the risk of type 2 diabetes. The genetic variability within transcription factors such as the SREBF1 gene, which are affected by fish oil intake may be the key to understanding the variability observed in the response of fasting insulin concentrations and insulin sensitivity[63]. SNPs within the SREBF1 gene have been frequently associated with type 2 diabetes or insulin resistance. A meta-analysis of four European genome screens found the strongest linkage with type 2 diabetes on chromosome 17p11.2-q22 where is located the SREBF1 gene[16]. One SNP (rs2297508) within SREBF1 gene has been reported across a few populations to be associated with the risk of type 2 diabetes[17, 18, 20, 21]. Felder et al.[21] found that the C/G and the G/G genotypes of rs2297508 had a ~1.4-fold higher risk of type 2 diabetes. In a French cohort, rs2297508 was also associated with obesity and type 2 diabetes independently of the obesity status[17]. Moreover, the SNP rs2297508 was related to sex-specific differences in the response of lipid and insulin concentrations as well as in HOMA-IR to a diet high in carbohydrates[64]. In their study, Zhang et al.[64] observed that the C allele of rs2297508 was associated with more favorable impacts on plasma triglyceride, fasting insulin and HOMA-IR than the G allele. In our study, the genotype alone was not associated with QUICKI values. However, a gene-diet interaction effect between rs2297508 and dietary PUFA intakes was observed. A large-scale gene-centric meta-analysis identified the SREBF1 gene as a type 2 diabetes loci among Europeans, with rs4925115 being the most significantly associated SNP[19]. The three tSNPs (rs12953299, rs4925115 and rs4925118) covered 100% of the known genetic variability within the SREBF1 gene. However, two of these tSNPs were in moderate LD (rs12953299 and rs4925115, r2 = 0.68) and were also in moderate to high LD with the most studied SREBF1 gene SNP rs2297508 (r2 = 0.62 and r2 = 0.95, respectively). Since SNPs within the SREBF1 gene including the promoter region, are in moderate to high LD, we cannot rule out the possibility that the tSNPs examined herein may also be in LD with SNPs within the SREBF1 gene promoter region which could affect its expression[65]. In the second cohort, the SNP rs2297508 from SREBF1 gene interacted with dietary PUFA intakes to affect insulin sensitivity (assessed by QUICKI). This SNP is located in exon 18c and is synonymous (Gly952Gly). Thus, these results may indicate that the genetic variability within the SREBF1 gene has an impact on the response of insulin sensitivity to n-3 PUFA and/or PUFA intakes.
Conclusion
To our knowledge this study was the first to examine associations between SNPs within the SREBF1 gene and the response of fasting insulin and insulin sensitivity to a fish oil supplementation. In this study, the genetic variability within the SREBF1 gene was associated with differences in the response of insulin and insulin sensitivity to a fish oil supplementation. SREBF1 gene may be an important candidate to study in order to understand the discrepancies observed in the impacts of fish oil on insulin resistance. Clinical trials taking into account the genetic variability within the SREBF1 gene and observing the impact of fish oil supplementation on insulin and insulin sensitivity are warranted. Moreover, the identification of individuals with a beneficial or adverse response to fish oil is important in order to appropriately recommend its supplementation.
Abbreviations
- CPT:
-
Cell preparation tube
- DHA:
-
Docosahexaenoic acid
- EPA:
-
Eicosapentaenoic acid
- FA:
-
Fatty acid
- FADS :
-
Fatty acid desaturase
- FFQ:
-
Food frequency questionnaire
- HOMA:
-
Homeostasis model assessment
- INAF:
-
Institute of Nutrition and Functional Foods
- IRS2 :
-
Insulin receptor substrate 2
- LD:
-
Linkage disequilibrium
- MAF:
-
Minor allele frequency
- n-3:
-
omega-3
- PBMC:
-
Peripheral blood mononuclear cell
- PL:
-
Plasma lipids
- PUFA:
-
Polyunsaturated fatty acid
- QUICKI:
-
Quantitative insulin sensitivity check index
- SFA:
-
Saturated fatty acid
- SNP:
-
Single nucleotide polymorphism
- SRE:
-
Sterol regulatory element
- SREBF1 :
-
Sterol regulatory element binding transcription factor 1
- Total-C:
-
Total-cholesterol
- UCP2 :
-
Uncoupling protein-2
References
Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F: SREBP transcription factors: master regulators of lipid homeostasis. Biochimie 2004, 86: 839-848. 10.1016/j.biochi.2004.09.018
Shimomura I, Shimano H, Horton JD, Goldstein JL, Brown MS: Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J Clin Invest 1997, 99: 838-845. 10.1172/JCI119247
Shimano H, Amemiya-Kudo M, Takahashi A, Kato T, Ishikawa M, Yamada N: Sterol regulatory element-binding protein-1c and pancreatic beta-cell dysfunction. Diabetes Obes Metab 2007, 9(Suppl 2):133-139.
Takahashi A, Motomura K, Kato T, Yoshikawa T, Nakagawa Y, Yahagi N, Sone H, Suzuki H, Toyoshima H, Yamada N, Shimano H: Transgenic mice overexpressing nuclear SREBP-1c in pancreatic beta-cells. Diabetes 2005, 54: 492-499. 10.2337/diabetes.54.2.492
Oh YS, Lee YJ, Kang Y, Han J, Lim OK, Jun HS: Exendin-4 inhibits glucolipotoxic ER stress in pancreatic beta cells via regulation of SREBP1c and C/EBPbeta transcription factors. J Endocrinol 2013, 216: 343-352. 10.1530/JOE-12-0311
Matsuzaka T, Shimano H, Yahagi N, Amemiya-Kudo M, Okazaki H, Tamura Y, Iizuka Y, Ohashi K, Tomita S, Sekiya M, Hasty A, Nakagawa Y, Sone H, Toyoshima H, Ishibashi S, Osuga J, Yamada N: Insulin-independent induction of sterol regulatory element-binding protein-1c expression in the livers of streptozotocin-treated mice. Diabetes 2004, 53: 560-569. 10.2337/diabetes.53.3.560
Lin J, Yang R, Tarr PT, Wu PH, Handschin C, Li S, Yang W, Pei L, Uldry M, Tontonoz P, Newgard CB, Spiegelman BM: Hyperlipidemic effects of dietary saturated fats mediated through PGC-1beta coactivation of SREBP. Cell 2005, 120: 261-273. 10.1016/j.cell.2004.11.043
Sekiya M, Yahagi N, Matsuzaka T, Najima Y, Nakakuki M, Nagai R, Ishibashi S, Osuga J, Yamada N, Shimano H: Polyunsaturated fatty acids ameliorate hepatic steatosis in obese mice by SREBP-1 suppression. Hepatology 2003, 38: 1529-1539. 10.1016/j.hep.2003.09.028
Hannah VC, Ou J, Luong A, Goldstein JL, Brown MS: Unsaturated fatty acids down-regulate srebp isoforms 1a and 1c by two mechanisms in HEK-293 cells. J Biol Chem 2001, 276: 4365-4372. 10.1074/jbc.M007273200
Bremer AA, Stanhope KL, Graham JL, Cummings BP, Ampah SB, Saville BR, Havel PJ: Fish oil supplementation ameliorates fructose-induced hypertriglyceridemia and insulin resistance in adult male rhesus macaques. J Nutr 2013, 144: 5-11.
Lanza IR, Blachnio-Zabielska A, Johnson ML, Schimke JM, Jakaitis DR, Lebrasseur NK, Jensen MD, Sreekumaran Nair K, Zabielski P: Influence of fish oil on skeletal muscle mitochondrial energetics and lipid metabolites during high-fat diet. Am J Physiol Endocrinol Metab 2013, 304: E1391-E1403. 10.1152/ajpendo.00584.2012
Gonzalez-Periz A, Horrillo R, Ferre N, Gronert K, Dong B, Moran-Salvador E, Titos E, Martinez-Clemente M, Lopez-Parra M, Arroyo V, Claria J: Obesity-induced insulin resistance and hepatic steatosis are alleviated by omega-3 fatty acids: a role for resolvins and protectins. FASEB J 2009, 23: 1946-1957. 10.1096/fj.08-125674
Wakutsu M, Tsunoda N, Shiba S, Muraki E, Kasono K: Peroxisome proliferator-activated receptors (PPARs)-independent functions of fish oil on glucose and lipid metabolism in diet-induced obese mice. Lipids Health Dis 2010, 9: 101. 10.1186/1476-511X-9-101
Tanaka N, Zhang X, Sugiyama E, Kono H, Horiuchi A, Nakajima T, Kanbe H, Tanaka E, Gonzalez FJ, Aoyama T: Eicosapentaenoic acid improves hepatic steatosis independent of PPARalpha activation through inhibition of SREBP-1 maturation in mice. Biochem Pharmacol 2010, 80: 1601-1612. 10.1016/j.bcp.2010.07.031
Kato T, Shimano H, Yamamoto T, Ishikawa M, Kumadaki S, Matsuzaka T, Nakagawa Y, Yahagi N, Nakakuki M, Hasty AH, Takeuchi Y, Kobayashi K, Takahashi A, Yatoh S, Suzuki H, Sone H, Yamada N: Palmitate impairs and eicosapentaenoate restores insulin secretion through regulation of SREBP-1c in pancreatic islets. Diabetes 2008, 57: 2382-2392. 10.2337/db06-1806
Demenais F, Kanninen T, Lindgren CM, Wiltshire S, Gaget S, Dandrieux C, Almgren P, Sjögren M, Hattersley A, Dina C, Tuomi T, McCarthy MI, Froguel P, Groop LC: A meta-analysis of four European genome screens (GIFT Consortium) shows evidence for a novel region on chromosome 17p11.2-q22 linked to type 2 diabetes. Hum Mol Genet 2003, 12: 1865-1873. 10.1093/hmg/ddg195
Eberle D, Clement K, Meyre D, Sahbatou M, Vaxillaire M, Le GA, Ferré P, Basdevant A, Froquel P, Foufelle F: SREBF-1 gene polymorphisms are associated with obesity and type 2 diabetes in French obese and diabetic cohorts. Diabetes 2004, 53: 2153-2157. 10.2337/diabetes.53.8.2153
Laudes M, Barroso I, Luan J, Soos MA, Yeo G, Meirhaeghe A, Logie L, Vidal-Puig A, Schafer AJ, Wareham NJ, O’Rahilly S: Genetic variants in human sterol regulatory element binding protein-1c in syndromes of severe insulin resistance and type 2 diabetes. Diabetes 2004, 53: 842-846. 10.2337/diabetes.53.3.842
Saxena R, Elbers CC, Guo Y, Peter I, Gaunt TR, Mega JL, Lanktree MB, Tare A, Castillo BA, Li YR, Johnson T, Bruinenberg M, Gilbert-Diamond D, Rajagopalan R, Voight BF, Balasubramanyam A, Barnard J, Bauer F, Baumert J, Bhangale T, Böhm BO, Braund PS, Burton PR, Chandrupatla HR, Clarke R, Cooper-DeHoff RM, Crook ED, Davey-Smith G, Day IN, de Boer A, et al.: Large-scale gene-centric meta-analysis across 39 studies identifies type 2 diabetes loci. Am J Hum Genet 2012, 90: 410-425. 10.1016/j.ajhg.2011.12.022
Grarup N, Stender-Petersen KL, Andersson EA, Jorgensen T, Borch-Johnsen K, Sandbaek A, Lauritzen T, Schmitz O, Hansen T, Pedersen O: Association of variants in the sterol regulatory element-binding factor 1 (SREBF1) gene with type 2 diabetes, glycemia, and insulin resistance: a study of 15, 734 Danish subjects. Diabetes 2008, 57: 1136-1142. 10.2337/db07-1534
Felder TK, Oberkofler H, Weitgasser R, Mackevics V, Krempler F, Paulweber B, Patsch W: The SREBF-1 locus is associated with type 2 diabetes and plasma adiponectin levels in a middle-aged Austrian population. Int J Obes (Lond) 2007, 31: 1099-1103. 10.1038/sj.ijo.0803505
Liu JX, Liu J, Li PQ, Xie XD, Guo Q, Tian LM, Ma XQ, Zhang JP, Liu J, Gao JY: Association of sterol regulatory element-binding protein-1c gene polymorphism with type 2 diabetes mellitus, insulin resistance and blood lipid levels in Chinese population. Diabetes Res Clin Pract 2008, 82: 42-47. 10.1016/j.diabres.2008.06.017
Ogawa S, Abe T, Nako K, Okamura M, Senda M, Sakamoto T, Ito S, DIMS Study Group: Eicosapentaenoic acid improves glycemic control in elderly bedridden patients with type 2 diabetes. Tohoku J Exp Med 2013, 231: 63-74. 10.1620/tjem.231.63
Lopez-Huertas E: The effect of EPA and DHA on metabolic syndrome patients: a systematic review of randomised controlled trials. Br J Nutr 2012, 107(Suppl 2):S185-S194.
Tierney AC, McMonagle J, Shaw DI, Gulseth HL, Helal O, Saris WH, Paniaqua JA, Golabek-Leszczynska I, Defoort C, Williams CM, Karsltröm B, Vessby B, Dembinska-Kiec A, Lopez-Miranda J, Blaak EE, Drevon CA, Gibney MJ, Lovegrove JA, Roche HM: Effects of dietary fat modification on insulin sensitivity and on other risk factors of the metabolic syndrome–LIPGENE: a European randomized dietary intervention study. Int J Obes (Lond) 2011, 35: 800-809. 10.1038/ijo.2010.209
Borkman M, Storlien LH, Pan DA, Jenkins AB, Chisholm DJ, Campbell LV: The relation between insulin sensitivity and the fatty-acid composition of skeletal-muscle phospholipids. N Engl J Med 1993, 328: 238-244. 10.1056/NEJM199301283280404
Jafari T, Fallah AA, Azadbakht L: Role of dietary n-3 polyunsaturated fatty acids in type 2 diabetes: a review of epidemiological and clinical studies. Maturitas 2013, 74: 303-308. 10.1016/j.maturitas.2013.01.008
Mori TA, Burke V, Puddey IB, Watts GF, O’Neal DN, Best JD, Beilin LJ: Purified eicosapentaenoic and docosahexaenoic acids have differential effects on serum lipids and lipoproteins, LDL particle size, glucose, and insulin in mildly hyperlipidemic men. Am J Clin Nutr 2000, 71: 1085-1094.
Woodman RJ, Mori TA, Burke V, Puddey IB, Watts GF, Beilin LJ: Effects of purified eicosapentaenoic and docosahexaenoic acids on glycemic control, blood pressure, and serum lipids in type 2 diabetic patients with treated hypertension. Am J Clin Nutr 2002, 76: 1007-1015.
Cormier H, Rudkowska I, Thifault E, Lemieux S, Couture P, Vohl MC: Polymorphisms in Fatty Acid Desaturase (FADS) gene cluster: effects on glycemic controls following an omega-3 Polyunsaturated Fatty Acids (PUFA) supplementation. Genes (Basel) 2013, 4: 485-498. 10.3390/genes4030485
Bouchard-Mercier A, Rudkowska I, Lemieux S, Couture P, Vohl MC: Polymorphisms, de novo lipogenesis, and plasma triglyceride response following fish oil supplementation. J Lipid Res 2013, 54: 2866-2873. 10.1194/jlr.M041590
Goulet J, Nadeau G, Lapointe A, Lamarche B, Lemieux S: Validity and reproducibility of an interviewer-administered food frequency questionnaire for healthy French-Canadian men and women. Nutr J 2004, 3: 13. 10.1186/1475-2891-3-13
McNamara JR, Schaefer EJ: Automated enzymatic standardized lipid analyses for plasma and lipoprotein fractions. Clin Chim Acta 1987, 166: 1-8. 10.1016/0009-8981(87)90188-4
Burstein M, Samaille J: [On a rapid determination of the cholesterol bound to the serum alpha- and beta-lipoproteins]. Clin Chim Acta 1960, 5: 609. 10.1016/0009-8981(60)90075-9
Albers JJ, Warnick GR, Wiebe D, King P, Steiner P, Smith L, Breckenridge C, Chow A, Kuba K, Weidman S, Arnett H, Wood P, Shlagenhaft A: Multi-laboratory comparison of three heparin-Mn2+ precipitation procedures for estimating cholesterol in high-density lipoprotein. Clin Chem 1978, 24: 853-856.
Friedewald WT, Levy RI, Fredrickson DS: Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem 1972, 18: 499-502.
Desbuquois B, Aurbach GD: Use of polyethylene glycol to separate free and antibody-bound peptide hormones in radioimmunoassays. J Clin Endocrinol Metab 1971, 33: 732-738. 10.1210/jcem-33-5-732
Richterich R, Dauwalder H: [Determination of plasma glucose by hexokinase-glucose-6-phosphate dehydrogenase method]. Schweiz Med Wochenschr 1971, 101: 615-618.
Katz A, Nambi SS, Mather K, Baron AD, Follmann DA, Sullivan G, Quon MJ: Quantitative insulin sensitivity check index: a simple, accurate method for assessing insulin sensitivity in humans. J Clin Endocrinol Metab 2000, 85: 2402-2410. 10.1210/jcem.85.7.6661
Antuna-Puente B, Faraj M, Karelis AD, Garrel D, Prud’homme D, Rabasa-Lhoret R, Bastard JP: HOMA or QUICKI: is it useful to test the reproducibility of formulas? Diabetes Metab 2008, 34: 294-296. 10.1016/j.diabet.2008.02.001
Muniyappa R, Lee S, Chen H, Quon MJ: Current approaches for assessing insulin sensitivity and resistance in vivo: advantages, limitations, and appropriate usage. Am J Physiol Endocrinol Metab 2008, 294: E15-E26.
Livak KJ: Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal 1999, 14: 143-149. 10.1016/S1050-3862(98)00019-9
Cormier H, Rudkowska I, Paradis AM, Thifault E, Garneau V, Lemieux S, Couture P, Vohl MC: Association between Polymorphisms in the Fatty Acid Desaturase Gene Cluster and the Plasma Triacylglycerol Response to an n-3 PUFA Supplementation. Nutrients 2012, 4: 1026-1041. 10.3390/nu4081026
Rudkowska I, Paradis AM, Thifault E, Julien P, Tchernof A, Couture P, Lemieux S, Barbier O, Vohl MC: Transcriptomic and metabolomic signatures of an n-3 polyunsaturated fatty acids supplementation in a normolipidemic/normocholesterolemic Caucasian population. J Nutr Biochem 2013, 24: 54-61. 10.1016/j.jnutbio.2012.01.016
Shaikh NA, Downar E: Time course of changes in porcine myocardial phospholipid levels during ischemia. A reassessment of the lysolipid hypothesis. Circ Res 1981, 49: 316-325. 10.1161/01.RES.49.2.316
Kroger E, Verreault R, Carmichael PH, Lindsay J, Julien P, Dewailly E, Ayotte P, Laurin D: Omega-3 fatty acids and risk of dementia: the Canadian Study of Health and Aging. Am J Clin Nutr 2009, 90: 184-192. 10.3945/ajcn.2008.26987
Liew CC, Ma J, Tang HC, Zheng R, Dempsey AA: The peripheral blood transcriptome dynamically reflects system wide biology: a potential diagnostic tool. J Lab Clin Med 2006, 147: 126-132. 10.1016/j.lab.2005.10.005
Rudkowska I, Raymond C, Ponton A, Jacques H, Lavigne C, Holub BJ, Marette A, Vohl MC: Validation of the use of peripheral blood mononuclear cells as surrogate model for skeletal muscle tissue in nutrigenomic studies. OMICS 2011, 15: 1-7. 10.1089/omi.2010.0073
Paradis AM, Godin G, Perusse L, Vohl MC: Associations between dietary patterns and obesity phenotypes. Int J Obes (Lond) 2009, 33: 1419-1426. 10.1038/ijo.2009.179
Gao X, Starmer J, Martin ER: A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol 2008, 32: 361-369. 10.1002/gepi.20310
Thifault E, Cormier H, Bouchard-Mercier A, Rudkowska I, Paradis AM, Garneau V, Ouellette C, Lemieux S, Couture P, Vohl MC: Effects of age, sex, body mass index and APOE genotype on cardiovascular biomarker response to an n-3 polyunsaturated fatty acid supplementation. J Nutrigenet Nutrigenomics 2013, 6: 73-82. 10.1159/000350744
National Cholesterol Education Program (NCEP) Expert Panel: Third Report of the National Cholesterol Education Program (NCEP) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (Adult Treatment Panel III) final report. Circulation 2002, 106: 3143-3421.
Canadian Diabetes Association: Screening for and Diagnosing Diabetes. [http://guidelines.diabetes.ca/ScreeningAndDiagnosis/Screening]
Hrebicek J, Janout V, Malincikova J, Horakova D, Cizek L: Detection of insulin resistance by simple quantitative insulin sensitivity check index QUICKI for epidemiological assessment and prevention. J Clin Endocrinol Metab 2002, 87: 144-147. 10.1210/jcem.87.1.8292
Ide T, Shimano H, Yahagi N, Matsuzaka T, Nakakuki M, Yamamoto T, Nakagawa Y, Takahashi A, Suzuki H, Sone H, Toyoshima H, Fukamizu A, Yamada N: SREBPs suppress IRS-2-mediated insulin signalling in the liver. Nat Cell Biol 2004, 6: 351-357. 10.1038/ncb1111
Medvedev AV, Robidoux J, Bai X, Cao W, Floering LM, Daniel KW, Collins S: Regulation of the uncoupling protein-2 gene in INS-1 beta-cells by oleic acid. J Biol Chem 2002, 277: 42639-42644. 10.1074/jbc.M208645200
Chan CB, MacDonald PE, Saleh MC, Johns DC, Marban E, Wheeler MB: Overexpression of uncoupling protein 2 inhibits glucose-stimulated insulin secretion from rat islets. Diabetes 1999, 48: 1482-1486. 10.2337/diabetes.48.7.1482
Price PT, Nelson CM, Clarke SD: Omega-3 polyunsaturated fatty acid regulation of gene expression. Curr Opin Lipidol 2000, 11: 3-7. 10.1097/00041433-200002000-00002
Wu JH, Micha R, Imamura F, Pan A, Biggs ML, Ajaz O, Djousse L, Hu FB, Mozaffarian D: Omega-3 fatty acids and incident type 2 diabetes: a systematic review and meta-analysis. Br J Nutr 2012, 107(Suppl 2):S214-S227.
Friedberg CE, Janssen MJ, Heine RJ, Grobbee DE: Fish oil and glycemic control in diabetes. A meta-analysis. Diabetes Care 1998, 21: 494-500. 10.2337/diacare.21.4.494
Puhakainen I, Ahola I, Yki-Jarvinen H: Dietary supplementation with n-3 fatty acids increases gluconeogenesis from glycerol but not hepatic glucose production in patients with non-insulin-dependent diabetes mellitus. Am J Clin Nutr 1995, 61: 121-126.
Kamolrat T, Gray SR, Thivierge MC: Fish oil positively regulates anabolic signalling alongside an increase in whole-body gluconeogenesis in ageing skeletal muscle. Eur J Nutr 2013, 52: 647-657. 10.1007/s00394-012-0368-7
Berger A, Mutch DM, German JB, Roberts MA: Dietary effects of arachidonate-rich fungal oil and fish oil on murine hepatic and hippocampal gene expression. Lipids Health Dis 2002, 1: 2. 10.1186/1476-511X-1-2
Zhang Z, Gong RR, Du J, Xiao LY, Duan W, Zhou XD, Fang DZ: Associations of the SREBP-1c gene polymorphism with gender-specific changes in serum lipids induced by a high-carbohydrate diet in healthy Chinese youth. Appl Physiol Nutr Metab 2011, 36: 226-232. 10.1139/h11-005
Nagata R, Nishio Y, Sekine O, Nagai Y, Maeno Y, Ugi S, Maegawa H, Kashiwagi A: Single nucleotide polymorphism (-468 Gly to A) at the promoter region of SREBP-1c associates with genetic defect of fructose-induced hepatic lipogenesis [corrected]. J Biol Chem 2004, 279: 29031-29042. 10.1074/jbc.M309449200
Acknowledgments
This research would not have been possible without the excellent collaboration of the participants. We would like to thank Hubert Cormier, Véronique Garneau, Alain Houde, Catherine Ouellette, Catherine Raymond, Élisabeth Thifault and the nurses, Danielle Aubin and Steeve Larouche, for their participation in the recruitment of the participants, the study coordination and the data collection.
This work was supported by a grant from CIHR - (MOP229488). ABM is supported by a studentship from the Fonds de recherche en santé du Quebec (FRQS) and by a Canadian Institutes of Health Research (CIHR) Frederick Banting and Charles Best Canada Graduate Scholarships Doctoral Awards (201210GSD-304012-190387) and MCV is Tier 1 Canada Research Chair in Genomics Applied to Nutrition and Health.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
IR, SL, PC, LP and MCV designed research; ABM conducted research with the research professionals; IR, SL, PC, LP and MCV provided essential reagents or provided essential materials; ABM analyzed data and performed statistical analysis; ABM wrote paper; ABM, IR, SL, PC, LP and MCV had primary responsibility for final content; All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Bouchard-Mercier, A., Rudkowska, I., Lemieux, S. et al. SREBF1 gene variations modulate insulin sensitivity in response to a fish oil supplementation. Lipids Health Dis 13, 152 (2014). https://doi.org/10.1186/1476-511X-13-152
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1476-511X-13-152