Abstract
Background
Serum amyloid A (SAA) is a kind of apolipoprotein. Several studies indicated that SAA genetic polymorphism rs12218 was associated with carotid atherosclerosis, peripheral arterial disease, and serum uric acid levels. However, the relation between rs12218 and lipid levels remains unclear. This study assessed the correlation between SAA1 gene rs12218 polymorphism and lipid levels in a Chinese population.
Methods
A total of 823 participants were selected from the subjects for health check in Shanghai Huashan hospital from Jan. 2013 to Mach. 2013. Correlations between rs12218 polymorphism and lipid levels were investigated through the identification of rs12218 genotypes using the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP).
Results
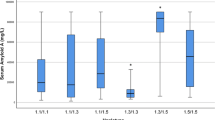
We found that the SNP rs12218 was associated with triglyceride (TG), total cholesterol (TC), and low-density lipoprotein (LDL-C) levels by analyses of a dominant model (P<0.001, P=0.002, P=0.003, respectively), a recessive model (P <0.001, P=0.001, P=0.005, respectively) and an additive model (P < 0.001, P=0.001, P=0.002, respectively), and the difference remained significant after the adjustment of sex, age, alcohol intake, and smoking (All P < 0.01).
Conclusion
Our results indicated that the rs12218 in the SAA1 gene was associated with lipid levels in a Chinese population.
Similar content being viewed by others
Background
Serum amyloid A (SAA) is a kind of apolipoprotein and is primarily synthesized in the liver by activated monocytes and macrophages [1]. As an apolipoprotein, SAA is associated with lipid level, such as high-density lipoprotein (HDL-C), and during inflammation can contribute up to 80% of its apoprotein composition [2].
The human SAA gene cluster on the short arm of chromosome 11, localized to band p15.1, contains four related genes, SAA1-4, within a 150-kb region [3]. Only the SAA1 and SAA2 genes encode acute-phase SAAs (SAA1 and SAA2 proteins), so a lot of research focuses on them. Recently, Carty et al. [4] reported an association of SAA1 and SAA2 gene polymorphisms and carotid intima-media thickness (cIMT), HDL-C, and total CVD. Many studies have demonstrated that rs12218 in the SAA1 gene was associated with carotid atherosclerosis [5] and peripheral arterial disease [6]. However, the relationships between SAA gene polymorphism and lipid level remain unclear.
In the present study, we aim to study the relationship between SAA1 gene polymorphism (rs12218) and lipids levels.
Results and discussion
This study consists of 823 subjects. The clinical and metabolic characteristics of the study population are shown in Table 1.
The distribution of genotype was in Hardy-Weinberg equilibrium (P>0.05, data not shown). Table 2 shows detailed information for rs12218 as well as the allele frequencies.
We found that the SNP rs12218 was associated with triglyceride (TG), total cholesterol (TC), and low-density lipoprotein (LDL-C) levels by analyses of a dominant model (P<0.001, P=0.002, P=0.003, respectively), recessive model (P <0.001, P=0.001, P=0.005, respectively) and additive model (P < 0.001, P=0.001, P=0.002, respectively), and the difference remained significant after the adjustment of sex, age, alcohol intake, and smoking (All P < 0.01) (Table 3).
In the present study, we found that variation in the SAA1 gene is associated with TG, TC, and LDL-C levels in Chinese population. In the early 1970s, SAA was identified as the plasma protein responsible for forming tissue deposits called “amyloid (AA-type)” seen in diseases with underlying persistent acute inflammation [7, 8]. Soon after its discovery, SAA was shown to be an acute phase protein produced by the liver within hours of tissue injury regardless of cause. Its plasma concentration can increase a 1000-fold within 24 h [9, 10]. In plasma, SAA is associated with HDL-C [11, 12] and, during severe inflammation, can contribute 80% of its apo-protein composition [13]. The displaced apoA-I is rapidly cleared by the liver and kidneys [14], together with a sharp decline in apoA-I gene expression during inflammation [15]. A relationship between the SAA1 gene polymorphism and cardiovascular diseases has been reported previously [1, 2, 4]. Previous studies have investigated the SAA1 rs12218 polymorphism in the Chinese population, but its relationship with lipid level has not been thoroughly investigated. Xie et al. [5] and Feng et al. [1] reported the relationship between rs12218 and lipid level, however, they did not reach to the same conclusion. Feng et al. reported an association between HDL-C concentration and rs12218, he found that in the osteoporosis group the rs12218 was significantly associated with plasma TC, HDL-C, and LDL-C levels (P=0.021, P=0.009, and P=0.009, respectively). However, this association was not found in the control group. And, they did not find the TG level was significantly associated with rs12218. Xie et al. also found rs12218 was significantly associated with HDL-C concentration, but they did not found the TG, LDL-C and TC was associated with rs12218. In the present study, we found rs12218 associated with TG, TC and LDL-C level both in the additive model, but also in dominant model and recessive model. After adjusted for some confounders the difference remains significant. This discrepancy may be result from the different population selection. And the lager-sample study related to this issue was expected.
Conclusions
In conclusion, the SAA1 gene polymorphism was associated with lipid level in a Chinese population.
Subjects and methods
Subjects
This study was approved by the Ethics Committee of the Shanghai Fudan University and was conducted according to the standards of the Declaration of Helsinki. Written informed consent was obtained from the participants. A total of 823 participants were selected from the subjects for health check in our hospital from Jan. 2013 to Mach. 2013 in Shanghai Huashan Hospital. These subjects were free from diabetes, hypertension, or any history of coronary artery disease (CAD). Height, body weight, and blood pressure were measured as described previously [16, 17]. Smoking and drinking status was self-reported by study questionnaire as described previously [17]. We measured the fasting plasma concentration of total cholesterol, triglyceride (TG), low-density lipoprotein (LDL), high-density lipoprotein (HDL) and glucose using an equipment for chemical analysis (Dimension AR/AVL Clinical Chemistry System, Newark, NJ) employed by the Clinical Laboratory Department of Shanghai Huashan hospital.
Rs12218 genotyping
Genomic DNA was extracted from the peripheral blood leukocytes using a DNA extraction Kit (Beijing Bioteke Co. Ltd, China). We genotyped rs12218 according to the protocol described previously [2]. To ensure the results were verified, 10% of the genotyped samples were duplicated, and at least one positive and one negative control per 96-well DNA plate were used in our assays. The accuracy of the genotyping was determined by assessing the genotype concordance between duplicate samples. We obtained a 100% concordance between the genotyped duplicate samples for the SNP. The genotyping success rate was 100%.
Statistical analysis
All analyses were carried out using SPSS version 17.0 (SPSS Inc., Chicago, IL, USA). The Hardy-Weinberg equilibrium was assessed using chi-square analysis. The characteristics of the study population were expressed as the mean ± standard deviation or as a ratio. Fasting triglycerides were log-transformed using natural logarithms for analysis. General linear model analysis was undertaken to test for associations between SNP genotypes and lipid levels after adjusting for confounding variables. Single-SNP effects with continuous variables were analyzed using linear regression using three models. These were the additive (common allele homozygotes coded as 1, heterozygotes as 2, and recessive allele homozygotes as 3); dominant (common allele homozygotes coded as 1 and heterozygotes and recessive allele homozygotesas 2); and recessive (common allele homozygotes and heterozygotes coded as1 and recessive allele homozygotes as 2) models as described previously [16]. Normality was assessed by plotting the residuals.
Abbreviations
- SAA:
-
Serum amyloid A
- CAD:
-
Coronary artery disease
- TG:
-
Triglycerides
- TC:
-
Total cholesterol
- HDL-C:
-
High-density lipoprotein
- LDL-C:
-
Low-density lipoprotein.
References
Feng ZP, Li XY, Jiang R, Deng HC, Yang M, Zhou Q, Que WJ, Du J: Associations of SAA1 gene polymorphism with lipid levels and osteoporosis in Chinese women. Lipids Health Dis. 2013, 12 (1): 39.
Xie X, Ma YT, Yang YN, Li XM, Fu ZY, Zheng YY, Ma X, Chen BD, Liu F, Huang Y, Yu ZX, Chen Y: Serum uric acid levels are associated with polymorphism in the SAA1 gene in Chinese subjects. PLoS One. 2012, 7 (6): e40263.
Moriguchi M, Terai C, Kaneko H, Koseki Y, Kajiyama H: A novel single-nucleotide polymorphism at the 59-flanking region of SAA1 associated with risk of type AA amyloidosis secondary to rheumatoid arthritis. Arth Rheum. 2001, 44: 1266-1272.
Carty CL, Heagerty P, Heckbert SR, Enquobahrie DA, Jarvik GP, Davis S, Tracy RP, Reiner AP: Association of genetic variation in serum amyloid-a with cardiovascular disease and interactions with IL6, IL1RN, IL1band TNF gene in the cardiovascular health study. J Ather Throm. 2009, 16: 419-430.
Xie X, Ma YT, Yang YN, Fu ZY, Li XM, Huang D, Ma X, Chen BD, Liu F: Polymorphisms in the SAA1/2 gene are associated with carotid intima media thickness in healthy Han Chinese subjects: the cardiovascular risk survey. PLoS One. 2010, 5 (11): e13997-
Xie X, Ma YT, Yang YN, Fu ZY, Li XM, Zheng YY, Huang D, Ma X, Chen BD, Liu F: Polymorphisms in the SAA1 gene are associated with ankle-to-brachial index in Han Chinese healthy subjects. Blood Press. 2011, 20 (4): 232-238.
Anthony D, Seow HJ, Uddin M, Thompson M, Dousha L, Vlahos R, Irving LB, Levy BD, Anderson GP, Bozinovski S: Serum amyloid a promotes lung neutrophilia by increasing IL-17A levels in the mucosa and γδ T cells. Am J Respir Crit Care Med. 2013, Epub ahead of print.
Moran G, Carcamo C, Concha M, Folch H: Expression of the protein serum amyloid a (SAA) in response to aspergillus fumigatus in murine models of allergic airway inflammation. Rev Iberoam Micol. 2013, Epub ahead of print.
Patke S, Srinivasan S, Maheshwari R, Srivastava SK, Aguilera JJ, Colón W, Kane RS: Characterization of the oligomerization and aggregation of human serum amyloid a. PLoS One. 2013, 8 (6): e64974. 10.1371/journal.pone.0064974. Print 2013.
Li B, Dong Z, Liu H, Xia YF, Liu XM, Luo BB, Wang WK, Li B, Gao F, Zhang C, Zhang MX, Zhang Y, An FS: Serum amyloid a stimulates lipoprotein-associated phospholipase A2 expression in vitro and in vivo. Atherosclerosis. 2013, 228 (2): 370-379. 10.1016/j.atherosclerosis.2013.03.023.
Kumon Y, Suehiro T, Ikeda Y, Yoshida K, Hashimoto K, Ohno F: Influence of serum amyloid a protein on high-density lipoprotein in chronic inflammatory disease. Clin Biochem. 1993, 26 (6): 505-511.
Wroblewski JM, Jahangiri A, Ji A, de Beer FC, van der Westhuyzen DR, Webb NR: Nascent HDL formation by hepatocytes is reduced by the concerted action of serum amyloid a and endothelial lipase. J Lipid Res. 2011, 52 (12): 2255-2261. 10.1194/jlr.M017681.
Kim SR, Kondo F, Otono Y, Imoto S, Ando K, Hirakawa M, Fukuda K, Sasaki M, Kim SK, Komaki T, Tsuchida S, Kobayashi S, Matsuoka T, Kudo M: Serum amyloid a and C-reactive protein positive nodule in alcoholic liver cirrhosis, hard to make definite diagnosis. Hepatol Res. 2013, Epub ahead of print.
Malle E, Leonhard B, Knipping G, Sattler W: Effects of cytokines, butyrate and dexamethasone on serum amyloid a and apolipoprotein a-I synthesis in human HUH-7 hepatoma cells. Scand J Immunol. 1999, 50 (2): 183-187.
Ancsin JB, Kisilevsky R: Laminin interactions with the apoproteins of acute-phase HDL: preliminary mapping of the laminin binding site on serum amyloid a. Amyloid. 1999, 6 (1): 37-47.
Xie X, Ma YT, Yang YN, Li XM, Liu F, Huang D, Fu ZY, Ma X, Chen BD, Huang Y: Alcohol consumption and ankle-to-brachial index: results from the cardiovascular risk survey. PLoS One. 2010, 5 (12): e15181-10.1371/journal.pone.0015181.
Xie X, Ma YT, Yang YN, Fu ZY, Ma X, Huang D, Li XM, Chen BD, Liu F, Huang Y, Liu C, Zhang XL, Zheng YY, Baituola G, Wang BZ, Du L, Gao X: Alcohol consumption and carotid atherosclerosis in china: the cardiovascular risk survey. Eur J Prev Cardiol. 2012, 19 (3): 314-321.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The author declare that they have no competing interests.
Authors’ contributions
XLX and XTS carried out the molecular genetic studies and drafted the manuscript. LWP and GQH carried out the genotyping. JCH, XTS and XLX participated in the design of the study and performed the statistical analysis. MS and YQW conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Xiao-Lin Xu, Xiao-Tian Sun contributed equally to this work.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Xu, XL., Sun, XT., Pang, L. et al. Rs12218 In SAA1 gene was associated with serum lipid levels. Lipids Health Dis 12, 116 (2013). https://doi.org/10.1186/1476-511X-12-116
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1476-511X-12-116