Abstract
Background
Pancreatic cancer is characterized by constitutive activation of mitogen-activated protein kinase (MAPK). Activation of MAPK is associated with the upregulation of genes implicated in the proliferation and survival of pancreatic cancer cells. We hypothesized that knockdown of these MAPK-associated molecules could produce notable anticancer phenotypes.
Methods
A RNA interference-mediated knockdown screening of 78 MAPK-associated molecules previously identified was performed to find molecules specifically associated with proliferation of pancreatic cancer cells in vitro. Expression of an identified molecule in pancreatic cancer tissues was examined by immunohistochemistry. In vivo tumorigenicity of cancer cells with stable knockdown of the molecule was assayed by using xenograft models. Flow cytometry and live cell imaging were employed to assess an association of the molecule with cell cycle.
Results
The knockdown screening revealed that knockdown of SON, the gene encoding SON, which is a large serine/arginine-rich protein involved in RNA processing, substantially suppressed pancreatic cancer cell proliferation and survival in vitro and tumorigenicity in vivo. SON expression was higher in ductal adenocarcinomas than in cells of normal ducts and precursor lesions in pancreatic cancer tissues. Knockdown of SON induced G2/M arrest and apoptosis in cultured cancer cells. The suppressive effect of SON knockdown on proliferation was less pronounced in cultured normal duct epithelial cells. SON formed nuclear speckles in the interphase of the cell cycle and dispersed in the cytoplasm during mitosis. Live cell imaging showed that SON diffusely dispersed in the early mitotic phase, accumulated in some foci in the cytoplasm in the late mitotic phase, and gradually reassembled into speckles after mitosis.
Conclusion
These results indicate that SON plays a critical role in the proliferation, survival, and tumorigenicity of pancreatic cancer cells, suggesting that SON is a novel therapeutic molecular target for pancreatic cancer.
Similar content being viewed by others
Background
Pancreatic cancer is a leading cause of cancer-related deaths [1, 2]. Despite advancements in diagnostic and therapeutic modalities, the 5-year survival rate of patients with pancreatic cancer is less than 10% [3]. This poor prognosis elicits an urgent need for the development of effective diagnostic and therapeutic measures to improve patient survival. Molecular medicine may be able to fulfill this need, as exemplified by imatinib in the treatment of chronic myeloid leukemia [4]. Pancreatic cancer is characterized by constitutive activation of mitogen-activated protein kinase (MAPK), due to gain-of-function mutations in KRAS or BRAF and loss-of-function of dual specificity phosphatase 6 (DUSP6) [5–7]. Active MAPK translocates to the nucleus, activates transcription factors, and induces the expression of a variety of genes [8]. In a previous study, we screened the genome for downstream targets of MAPK and identified 78 molecules specifically associated with MAPK activity in pancreatic cancer cells [9]. These MAPK-associated molecules include molecules implicated in DNA replication, RNA editing, spindle formation, mitosis, signal transduction, and membrane trafficking. These biological processes play critical roles in the survival, maintenance, and proliferation of pancreatic cancer cells. We hypothesized that molecular targeting of these MAPK-associated molecules could result in notable anticancer phenotypes, as we previously observed by targeting AURKA[9, 10]. In this study, we performed a systematic knockdown screening of MAPK-associated molecules in pancreatic cancer cells.
Results
Knockdown screening of MAPK-modulated genes in pancreatic cancer cells
We performed knockdown screening using a pancreatic cancer cell line, MIA PaCa-2, and custom-designed short interfering RNAs (siRNAs) targeting all the 78 MAPK-modulated genes that were previously identified and isolated in the cell line (Additional file 1: Table S1) [9]. The cells were transiently transfected with each of the 78 siRNAs, and in vitro proliferation was subsequently examined for 5 consecutive days. This screening showed that proliferation of cancer cells was suppressed to variable degrees depending on the individual gene targeted (Figure 1). Knockdown of AURKB, CENPA, EBNA1BP2, GOLT1A, KIF11, NEDD4L, SON, TPX2, or WDR5 suppressed proliferation by more than 50% compared with control. Among these targets, we focused on SON for further study because it showed the most substantial suppressive effect. This gene encodes a nuclear speckle protein, SON, which is involved in RNA processing.
Knockdown screening of MAPK-associated genes in pancreatic cancer cells. Proliferation of MIA PaCa-2, a pancreatic cancer cell line, transfected with siRNA targeting various genes associated with MAPK (indicated on the horizontal axis), was determined by MTT assay on day 5 post-transfection. Plotted values are expressed relative to cells infected with control siRNA to a nonspecific sequence (Nonsp). Plots represent an average of 2 independent experiments; each experiment includes data from 8 independent transfection wells. Knockdown of SON (closed column) showed the most remarkable anti-proliferative phenotype.
Knockdown of SON attenuates proliferation in vitro, considerably in pancreatic cancer cells but less remarkably in normal phenotype cells
The in vitro suppressive effect of siRNA targeting SON on proliferation was reanalyzed in detail by using MIA PaCa2; PCI-35, a pancreatic cancer cell line with an aggressive phenotype; and HPDE, an immortalized normal human pancreatic duct epithelial cell line [7, 11–13]. The suppressive effects of SON knockdown on cell proliferation appeared to be fatal in MIA PaCa-2, static in PCI-35, and insignificant in HPDE (Figure 2A). The effects of siRNA on SON expression were assayed by an immunoblotting method, which showed 77%, 10%, and 48% reduction of SON expression in MIA PaCa-2, PCI-35, and HPDE, respectively (Figure 2B). These results indicated that SON knockdown attenuated the in vitro proliferation of pancreatic cancer cells. The attenuation of proliferation depended on the efficiency of SON knockdown in pancreatic cancer cells, but was less remarkably affected in normal phenotype cells.
A. Proliferation of pancreatic cancer cells (MIA PaCa-2 and PCI-35) and normally phenotypic duct epithelial cells (HPDE) transfected with siRNA against SON (siSON) or a nonspecific sequence (siNons) and measured by MTT assay. The plots represent an average of 2 independent experiments; experiment includes data from 8 independent transfection wells. B. Expression of SON in cells transfected with siSON or siNons is shown in immunoblots probed with anti-SON antibody (SON) or anti-beta actin antibody (ACTB). C. Colony formation assay of pancreatic cancer cells transfected with vectors expressing shRNA targeting SON (shSON) or a non-specific sequence (shNons).
Stable knockdown of SON reduces the survival of pancreatic cancer cells in vitro
We next constructed a vector expressing short hairpin RNA (shRNA) identical to the SON siRNA when processed. We examined the effect of stable knockdown of SON on the survival of pancreatic cancer cells in vitro using a colony formation assay. We found that stable knockdown of SON strongly attenuated the survival of cancer cells, even in PCI-35 cells, in which transient transfection of siRNA targeting SON modestly suppressed proliferation (Figure 2C).
SON is overexpressed in pancreatic ductal adenocarcinomas
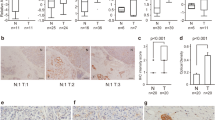
To establish the native expression of SON in pancreatic cancer, we examined 34 tissues with pancreatic ductal adenocarcinoma that were surgically resected. Immunohistochemistry showed that SON was strongly expressed in the nuclei of cancer cells in most ductal adenocarcinomas significantly more obviously than in the nuclei of non-neoplastic ducts or pancreatic intraepithelial neoplasia (PanIN), a precursor lesion of ductal adenocarcinoma (p < 0.001 by ANOVA) (Figure 3 and Table 1). This result indicates that SON is specifically overexpressed in pancreatic cancer.
Immunohistochemical examination of SON expression in pancreatic cancer tissues. Diaminobenzidine and hematoxylin was used as a chromogen and a counter stain, respectively. A and B. Ductal adenocarcinomas (ducts labeled in C) strongly express SON in nuclei, more obviously than normal ductal cells (ducts labeled in N). The normal duct in panel B was partially (lower half) involved with carcinoma cells (original magnification, 100×). C. A high-powered view of ductal adenocarcinoma shows strong expression of SON in nuclei (original magnification, 200×). D. Pancreatic intraepithelial neoplasia, a precursor lesion of ductal adenocarcinoma, shows less obvious expression of SON (original magnification, 100×).
Knockdown of SON retards the tumorigenicity of pancreatic cancer cells in vivo
We then performed a tumorigenicity assay using stably transfected pancreatic cancer cell clones carrying the shRNA vector targeting SON. Several stably transfected clones of MIA PaCa-2 and PCI-35 cells were obtained, and expression of SON was determined by real-time quantitative PCR. SON expression was lowest, reduced by 50%, in an MIA PaCa-2 clone (Figure 2D). We could not obtain any stably transfected PCI-35 clones in which SON expression was obviously reduced. This was probably because PCI-35, unlike MIA PaCa-2, could not survive modest knockdown of SON, which strongly suppresses the survival of cancer cells in vitro. The stably transfected clone of MIA PaCa-2 was inoculated into the subcutis of nude mice, and tumorigenicity was monitored. After 4 weeks, tumorigenicity was significantly retarded (Figure 4A).
A. Left panel: Expression of SON in cloned cells (MIA PaCa-2) stably transfected with shSON or shNons. Middle panel: An example image of a nude mouse with a xenograft of MIA PaCa-2 clones stably transfected with shSON or shNons at 4 weeks after inoculation in the subcutis. Right panel: Average growth of tumors from xenografts of MIA PaCa-2 cells stably transfected with shSON or shNons in the subcutis of 4 nude mice. One (*) and 2 (**) asterisks indicate p < 0.05 and p < 0.01, respectively. Error bars denote 1 value of standard error. B. Cell cycle fractions of pancreatic cancer MIA PaCa-2 and PCI-35 cells transfected with short interfering RNA (siRNA) against SON (siSON) or a nonspecific sequence (siNons) as determined by flow cytometry.
Knockdown of SON induces cell cycle arrest and apoptosis
To determine the mechanism by which SON knockdown suppresses the proliferation and survival of pancreatic cancer cells, the DNA content of siRNA-transfected MIA PaCa-2 and PCI-35 cells was measured by flow cytometry, and the cell cycle was assessed. Knockdown of SON increased the fraction of cells in G2/M and sub-G1, indicating that the cells were in G2/M arrest and apoptosis (Figure 4B).
SON shuttles between the nucleus and cytoplasm depending on the cell cycle
To investigate the dynamics of intracellular SON expression and its role in mitosis, a vector expressing SON, tagged with enhanced green fluorescence protein (EGFP) at the amino terminus (EGFP-SON), was constructed and transfected into 293 cells. The dynamics of intracellular SON expression were then analyzed. Expression of EGFP-SON was confirmed by immunoblotting by using specific antibodies against SON or EGFP (Figure 5A). Confocal laser scanning images showed that EGFP-SON was expressed as speckles in the nuclei of cells in the interphase and was dispersed in the cytoplasm of cells in the mitotic phase (Figure 5B). Time-lapse live imaging of cells expressing EGFP-SON showed that SON dispersed diffusely in the cytoplasm in metaphase and anaphase, accumulated in some foci in the cytoplasm during telophase and cytokinesis, and gradually reassembled in nuclear speckles after cytokinesis as foci in the cytoplasm faded (Figure 5C). From metaphase, the reassembly into nuclear speckles took approximately 2 hours. These results indicate that SON shuttles between the nucleus and the cytoplasm depending on the phase of the cell cycle, transitioning from nuclear speckles and through diffuse dispersion and subsequent temporal accumulation in the cytoplasm, to slow reassembly into nuclear speckles during mitosis and the early G1 phase.
A. Immunoblot of total cell lysate from 293 cells transfected with pEGFP vector or pEGFP-SON probed with antibodies against EGFP or SON. B. Fluorescence images of 293 cells transfected with pEGFP-SON or pEGFP-vector. Note that EGFP-SON was speckled in the nuclei of interphase cells and diffusely dispersed in mitotic cells. Original magnification, 600×. C. Time-lapse images of 293 cells transfected with EGFP-SON. Sequential images taken at 10-minute intervals are shown (the number on each image indicates minutes from starting). EGFP-labeled SON dispersed in the cytoplasm during metaphase and anaphase (panels at 0 and 10 minutes), accumulated in small foci in the cytoplasm during telophase and cytokinesis (panels between 20 and 50 minutes), and then gradually reassembled in nuclear speckles (panels at 60 minutes and later). Original magnification, 400×.
Discussion
In this study, among many genes associated with MAPK, we found that knockdown of SON remarkably suppressed the proliferation, survival, and tumor formation of pancreatic cancer cells. The suppressive effect was less pronounced in normally phenotypic ductal cells. In primary pancreatic cancer tissues, SON was overexpressed in ductal adenocarcinomas compared with normal duct cells and PanINs. Knockdown of SON induced G2/M arrest and apoptosis. SON shuttled between the nucleus and cytoplasm depending on the phase of cell cycle. These results indicate that SON plays a crucial role in the proliferation, survival, and tumorigenicity of pancreatic cancer cells, thus suggesting that this molecule could be a prime therapeutic molecular target for pancreatic cancer.
Our investigation showed that knockdown of MAPK-associated molecules suppressed the proliferation of pancreatic cancer cells in vitro to variable degrees. We found that knockdown of AURKB, CENPA, EBNA1BP2, GOLT1A, KIF11, NEDD4L, SON, TPX2, or WDR5 strongly suppressed the proliferation. AURKB encodes aurora kinase B (AURKB), which is involved in chromosome segregation and cytokinesis during mitosis [14]. CENPA encodes centromere protein A (CENPA), which, by functioning as a replacement for histone H3 in centromeric nucleosomes, plays an essential role in kinetochore formation and functions in cellular mitosis [15]. EBNA1BP2 encodes a ribonucleoprotein, Epstein-Barr virus nuclear antigen 1-binding protein 2 (EBNA1BP2), which serves as a scaffold for ribosome biogenesis [16]. GOLT1A encodes Golgi transport 1A (GOLT1A), which functions as a transporter on the Golgi membrane [17]. KIF11 encodes a microtubule-dependent motor protein, kinesin family member 11 (KIF11), which plays a critical role in chromosome positioning during mitosis [18]. NEDD4L encodes neural precursor cell expressed, developmentally down-regulated 4-like, an E3 ubiquitin protein ligase (NEDD4L) that plays a role in polyubiquitination and proteasomal destruction of SMAD2/3 [19]. TPX2 encodes a homologue of Tpx2 of Xenopus (TPX2), a binding partner of aurora kinase A (AURKA) that plays a role in microtubule spindle formation [20]. WDR5 encodes WD repeat domain 5 (WDR5), which binds methylated histone H3 lysine 4 (H3K4) and is required for recruiting H3K4 methyltransferase [21]. Among these, AURKB, CENPA, KIF11, and TPX2 are involved in functions of the microtubule spindles and kinetochores, which are considered essential for cell mitosis. Because we screened by assaying the effects of knockdown of the MAPK-associated genes on in vitro proliferation of pancreatic cancer cells, molecules associated with the microtubules and kinetochores might be selectively represented in our screening. Interestingly, these microtubule kinetochore-associated molecules have already been studied as molecular targets in various cancers [22–25]. Nevertheless, of these MAPK-associated molecules, we found that knockdown of SON most remarkably suppressed proliferation, which led us to investigate SON in detail as a candidate molecular target.
SON encodes SON, a large protein harboring a serine or arginine-rich domain. It was first cloned as a gene encoding a protein with DNA-binding activity. However, subsequently, it turned out to be a nuclear speckle protein involved in RNA processing and required for proper and efficient splicing of pre-mRNAs [26–30]. In our study, knockdown of SON attenuated the proliferation, survival, and tumorigenicity of pancreatic cancer cells. These suppressive effects were attributable to cell cycle arrest at the G2/M phase and apoptosis induced by depletion of SON. The association between the depletion of SON and G2/M arrest has been reported to be associated with impairment of spindle pole separation, microtubule dynamics, and genome integrity due to inadequate RNA splicing of a specific set of cell cycle-related genes with weak splice sites, i.e., splice sites without the conserved sequence [30].
Pancreatic cancer cells were more susceptible to depletion of SON than normally phenotypic cells. This may be due to rapid progression through the cell cycle in cancer cells, which results in exaggerated dependence on SON to maintain efficient RNA processing of the cell cycle-related genes. This interpretation could be endorsed by the overexpression of SON we found in most ductal adenocarcinomas, compared with normal ductal cells or precursor lesions, which suggests that adenocarcinoma cells depend on SON more strongly than normal ductal cells and precursor lesions to maintain their phenotypes. These results suggest that depletion of SON may specifically lead to an anticancer phenotype. SON overexpression is purportedly due to the constitutive activation of MAPK in ductal adenocarcinoma; however, other possible causes, such as gene amplification or aberrations in protein turnover, cannot be ruled out and will be a subject of further study.
The dynamics of SON distribution during the cell cycle is not well known. We performed live-cell imaging of cells expressing EGFP-SON and observed that SON dispersed in the cytoplasm during early mitotic phase formed small foci in the cytoplasm in the late mitotic phase, and gradually redistributed as speckles in the nucleus as foci in the cytoplasm faded. The cytoplasmic small foci are supposed to be mitotic interchromatin granules that correspond to accumulations of nuclear speckle proteins in the cytoplasm in the late mitotic phase [31, 32]. These dynamics seem similar to the dynamics of another speckle protein, SF2, and are consistent with the idea that SON plays a role in the appropriate organization of RNA splicing factors [29, 33, 34].
The knockdown of SON by RNA interference showed sufficient anti-cancer phenotypes experimentally. For the RNA interference, vector-mediated stable transduction appeared to be more effective than oligonucleotide-based transient transduction as shown in Figure 2. Although the stable knockdown of SON by RNA interference could be an efficient molecular therapy for pancreatic cancer, the lack of a conventional method for tissue-specific, stable delivery of short, double-stranded RNA could limit the use of this approach in clinical therapeutics. Indeed, the use of RNA interference in clinical practice is generally not warranted. Recently, however, systemic delivery of siRNA combined with a special nanoparticle successfully knocked down a target gene in melanoma in a clinical trial [35]. The use of such a technique to attempt specific knockdown of SON in pancreatic cancer cells in a clinical model is worth trying and is an issue to be resolved in a future study. The results of this study also suggest that development of a molecule-oriented chemical substance against SON as therapy for pancreatic cancer is warranted.
Conclusion
This study indicates that SON is overexpressed and plays a critical role in the proliferation, survival, and tumorigenicity of pancreatic cancer cells, suggesting that SON is a novel therapeutic molecular target for pancreatic cancer.
Methods
Cell culture
Human pancreatic cancer cell lines, MIA PaCa-2 and PCI-35, and the human embryonic kidney cell line 293 were obtained and cultured as previously described [7, 9]. The immortalized human pancreatic duct-epithelial cell line, HPDE, was kindly provided by Dr. MS Tsao (Princess Margaret Hospital and Ontario Cancer Institute, Toronto, ON) and cultured as previously described [12].
Transfection of siRNA and cell proliferation assay
siRNAs targeting each downstream MAPK-associated molecule were custom designed and manufactured (RNAi Co. Ltd., Tokyo, Japan) (Additional file 1: Table S1). Cells were seeded at 5 × 103 cells/well in 96-well plates with 100 μL of appropriate culture medium and incubated at 37°C with 5% CO2 for 24 hours. Then, the medium was replaced with OPTI-MEM (Life Technologies, Carlsbad, CA), and the cells were transfected with siRNA at 10 nM with Oligofectamine (Life Technologies) according to the manufacturer’s recommendations. After 4 hours of incubation, the transfection reagent was replaced with the appropriate culture medium. A colorimetric cell proliferation assay—3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) assay—was performed daily for 5 days as previously described [7].
Colony formation assay with shRNA vectors
pSUPER vector (Oligoengine, Seattle, WA) was used for the construction of vectors expressing shRNAs by cloning the oligonucleotides 5′-GATCCCCGCATCTAGACGTTCTATGATTCAAGAGATCATAGAACGTCTAGATGCTTTTTA-3′ and 5′-AGCTTAAAAAGCATCTAGACGTTCTATGATCTCTTGAATCATAGAACGTCTAGATGCGGG-3′ to target SON (shRNA-SON), and 5′-GATCCCCGTACCGCACGTCATTCGTATTCAAGAGATACGAATGACGTGCGGTACTTTTTA-3′ and 5′-AGCTTAAAAAGTACCGCACGTCATTCGTATCTCTTGAATACGAATGACGTGCGGTACGGG-3′ to serve as a control harboring a nonspecific sequence against the human genome (shRNA-Nons) according to the manufacturer’s instructions. MIA PaCa-2 and PCI-35 cells were seeded at 1 × 105 cells/well in 6-well plates and incubated for 24 hours at 37°C with 5% CO2. The shRNA-SON vector or shRNA-Nons vector were transfected into the cells with LipofectamineTM reagent (Life Technologies) according to the manufacturer’s recommendations. The cells were dissociated with trypsin 48 hours after transfection and reseeded in three 10-cm tissue-culture dishes, containing the appropriate culture medium supplemented with 10% FBS and G418 (Life Technologies) at 400 μg/mL for PCI-35 and 500 μg/mL for MIA PaCa-2. After 3 weeks, the cells were fixed with 10% formalin solution and stained with hematoxylin. The number of colonies was assessed with the COLONY program (Fujifilm Co. Ltd., Tokyo, Japan).
Immunohistochemistry
Thirty-four formalin-fixed, paraffin-embedded tissues of pancreatic ductal adenocarcinoma that were surgically resected during 2006 and 2007 at Tokyo Women’s Medical University Hospital were studied. Indirect immunohistochemical staining was performed as previously described [36] by using a polyclonal anti-SON antibody (1:1200 dilution, Sigma, St. Louis, MO), a secondary antibody against rabbit immunoglobulin (Nichirei, Tokyo, Japan), and streptavidin solution (Nichirei). Use of the archival pathological tissues was approved by the ethics committee of Tokyo Women’s Medical University. Immunohistochemical results were evaluated among ductal lesions classified into adenocarcinoma, PanIN, or normal duct by scoring intensities of staining into 1, weak; 2, moderate; and 3, strong by comparing with normal ductal cells that showed weak staining or acinar cells that showed moderate staining. The scores were statistically analyzed by ANOVA by using PASW Statistics software (IBM Japan, Tokyo, Japan).
Quantitative real-time polymerase chain reaction assay
The TaqMan Gene Expression Assay and a 7500 Real-time PCR system (Life Technologies) were used to analyze the transcriptional expression of SON by using the absolute quantitative assay according to the manufacturer’s instructions. The expression of SON was assessed relative to the endogenous expression of GAPDH.
In vivo tumorigenicity assay
Pancreatic cancer cells stably transfected with shRNA vectors were isolated by cloning the surviving cells from the colony formation assay. These clones, in 50% matrigel/culture medium without FBS, were inoculated into the subcutis of BALB/c nude mice (Clea Japan Inc., Tokyo, Japan). Tumorigenicity was monitored weekly, and the tumor volume was calculated using the following formula: V = D × d2 × 0.4 (V, tumor volume; D, largest dimension; d, smallest dimension).
Flow cytometry
Flow cytometric analyses for cell cycle and apoptosis were performed as previously described [7].
Construction of the EGFP-SON vector
An expression vector containing the full coding sequence of SON cDNA (NM_138927) was constructed by assembling amplified products using KOD Plus DNA Polymerase and its specific buffer (TOYOBO, Osaka, Japan), appropriate paired primers, and pooled cDNA obtained from a fetal brain cDNA library (Stratagene/Agilent Technologies Inc., Santa Clara, CA) as follows. Paired primers used for amplification of cDNA fragments were C51, 5′-TTTAAGCTTATGGCGACCAACATCGAGCAG-3′ (melting temperature [Tm], 58°C) and C12, 5′-TAAGGGTGTTCTTGATCGCC-3′ (Tm, 52°C); C7, 5′-AGCCGCCGGAGAAGATCAAGG-3′ (Tm, 59°C) and C10, 5′-CAGGCTCTGAGGGCAAATTG-3′ (Tm, 53°C); and C5, 5′-TAAACTCAGTGAACCCAAACC-3′ (Tm, 50°C) and C52, 5′-TTTGGTACCTCAATACCTATTCAAGAAAAACATAC-3′ (Tm 48°C). Products amplified by PCR were sequentially cloned into the pFLAG-CMV-4 vector (Sigma, St. Louis, MO) at Hin dIII-Eco RI-Kpn I sites to obtain pFLAG-SON. The pEGFP-C2 vector (Clontech, Mountain View, CA) was modified by fill-in of its Xho I site to adjust the reading frame. The coding region of SON cDNA was prepared from pFLAG-SON by digestion with Hin dIII and Kpn I for the 3′ fragment and Hin dIII for the 5′ fragment. These fragments were sequentially cloned into the modified pEGFP-C2 vector at Hin dIII and Kpn I sites to obtain the pEGFP-SON vector. DNA sequences were confirmed by using BigDye® Terminator and a 3130x Genetic analyzer (Life Technologies).
Immunoblot
Denatured total cell lysate was separated in a 5–15% polyacrylamide gel and blotted onto a polyvinylidene fluoride membrane by using an XV Pantera MP System (DRC Co., Ltd. Tokyo, Japan) according to the manufacturer’s recommendations. The blotted membrane was probed with anti-SON antibody (Sigma), anti-beta actin antibody (Sigma), or anti-EGFP antibody (Clontech). Horseradish peroxidase-conjugated anti-rabbit or anti-mouse immunoglobulin antibodies (GE Healthcare UK Ltd., Buckinghamshire, UK) were used for the secondary antibody reaction. Blocking conditions and concentrations of antibodies were determined according to the manufacturers’ recommendations. Signals were visualized by reaction with ECL Detection Reagent (GE Healthcare UK Ltd.) and captured digitally by using an LAS 4000 Mini (Fujifilm Co. Ltd.) or by autoradiography. Intensities of bands were measured digitally using Image Gauge software (Fujifilm Co. Ltd.).
Laser scanning fluorescence imaging
The pEGFP-SON vector was transfected into 293 cells using Lipofectamine Plus (Life Technologies) according to the manufacturer’s recommendations. The transfected cells were incubated with Eagle’s Minimum Essential Medium (Sigma) supplemented with 10% FBS and 400 μg/mL G418. Stably transfected clones were obtained by cloning surviving cells using a cylinder cup. The isolated clones were seeded in a glass-bottom dish and incubated for 24 hours. The cells were incubated with a medium supplemented with 0.1 μg/mL Hoechst 33342 (Life Technologies) for 30 minutes. The medium was then replaced with fresh growth medium and examined under a confocal laser scanning microscope (LSM5, Carl-Zeiss Microimaging GmbH, Goettingen, Germany). Time-lapse images were obtained for 2 layers at 0- and 5-μm depth with 10-minute intervals over a total of 230 minutes.
Statistics
Student’s t-test was applied to analyze statistical differences using Statview 5.0 software (SAS Institute Inc., Cary, NC, USA). P values of <0.05 were considered statistically significant.
Abbreviations
- AURKA:
-
Aurora kinase A
- AURKB:
-
Aurora kinase B
- CENPA:
-
Centromere protein A
- DUSP6:
-
Dual specificity phosphatase 6
- EBNA1BP2:
-
Epstein-Barr virus nuclear antigen 1-binding protein 2
- EGFP:
-
Enhanced green fluorescence protein
- GOLT1A:
-
Golgi transport 1A
- H3K4:
-
Histone H3 lysine 4
- KIF11:
-
Kinesin family member 11
- MAPK:
-
Mitogen-activated protein kinase
- MTT:
-
3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide
- NEDD4L:
-
Neural precursor cell expressed, developmentally down-regulated 4-like, an E3 ubiquitin protein ligase
- Nons:
-
Non-specific sequence
- PanIN:
-
Pancreatic intraepithelial neoplasia
- shRNA:
-
Short hairpin RNA
- siRNA:
-
Short interfering RNA
- TPX2:
-
A homologue of Tpx2 of Xenopus
- WDR5:
-
WD repeat domain 5.
References
Cancer statistics in Japan. Edited by: Kato H, Sobue T, Katanoda K, Saito Y, Tsukuma H, Sakura N, Kitai A. 2008, National Cancer Center, Tokyo.
Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 2009, 59 (4): 225-249. 10.3322/caac.20006
Berrino F, De Angelis R, Sant M, Rosso S, Bielska-Lasota M, Coebergh JW, Santaquilani M: Survival for eight major cancers and all cancers combined for European adults diagnosed in 1995–99: results of the EUROCARE-4 study. Lancet Oncol. 2007, 8 (9): 773-783. 10.1016/S1470-2045(07)70245-0
O’Brien SG, Guilhot F, Larson RA, Gathmann I, Baccarani M, Cervantes F, Cornelissen JJ, Fischer T, Hochhaus A, Hughes T: Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2003, 348 (11): 994-1004. 10.1056/NEJMoa022457
Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M: Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988, 53 (4): 549-554.
Calhoun ES, Jones JB, Ashfaq R, Adsay V, Baker SJ, Valentine V, Hempen PM, Hilgers W, Yeo CJ, Hruban RH: BRAF and FBXW7 (CDC4, FBW7, AGO, SEL10) mutations in distinct subsets of pancreatic cancer: potential therapeutic targets. Am J Pathol. 2003, 163 (4): 1255-1260. 10.1016/S0002-9440(10)63485-2
Furukawa T, Sunamura M, Motoi F, Matsuno S, Horii A: Potential tumor suppressive pathway involving DUSP6/MKP-3 in pancreatic cancer. Am J Pathol. 2003, 162 (6): 1807-1815. 10.1016/S0002-9440(10)64315-5
Brunet A, Roux D, Lenormand P, Dowd S, Keyse S, Pouyssegur J: Nuclear translocation of p42/p44 mitogen-activated protein kinase is required for growth factor-induced gene expression and cell cycle entry. EMBO J. 1999, 18 (3): 664-674. 10.1093/emboj/18.3.664
Furukawa T, Kanai N, Shiwaku HO, Soga N, Uehara A, Horii A: AURKA is one of the downstream targets of MAPK1/ERK2 in pancreatic cancer. Oncogene. 2006, 25 (35): 4831-4839. 10.1038/sj.onc.1209494
Hata T, Furukawa T, Sunamura M, Egawa S, Motoi F, Ohmura N, Marumoto T, Saya H, Horii A: RNA interference targeting aurora kinase a suppresses tumor growth and enhances the taxane chemosensitivity in human pancreatic cancer cells. Cancer Res. 2005, 65 (7): 2899-2905. 10.1158/0008-5472.CAN-04-3981
Sun C, Yamato T, Furukawa T, Ohnishi Y, Kijima H, Horii A: Characterization of the mutations of the K-ras, p53, p16, and SMAD4 genes in 15 human pancreatic cancer cell lines. Oncol Rep. 2001, 8 (1): 89-92.
Furukawa T, Duguid WP, Rosenberg L, Viallet J, Galloway DA, Tsao MS: Long-term culture and immortalization of epithelial cells from normal adult human pancreatic ducts transfected by the E6E7 gene of human papilloma virus 16. Am J Pathol. 1996, 148 (6): 1763-1770.
Yamanaka S, Sunamura M, Furukawa T, Sun L, Lefter LP, Abe T, Yatsuoka T, Fujimura H, Shibuya E, Kotobuki N: Chromosome 12, frequently deleted in human pancreatic cancer, may encode a tumor-suppressor gene that suppresses angiogenesis. Lab Invest. 2004, 84 (10): 1339-1351. 10.1038/labinvest.3700160
Lens SM, Voest EE, Medema RH: Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer. 2010, 10 (12): 825-841. 10.1038/nrc2964
Verdaasdonk JS, Bloom K: Centromeres: unique chromatin structures that drive chromosome segregation. Nat Rev Mol Cell Biol. 2011, 12 (5): 320-332. 10.1038/nrm3107
Hirano Y, Ishii K, Kumeta M, Furukawa K, Takeyasu K, Horigome T: Proteomic and targeted analytical identification of BXDC1 and EBNA1BP2 as dynamic scaffold proteins in the nucleolus. Genes Cells. 2009, 14 (2): 155-166. 10.1111/j.1365-2443.2008.01262.x
Conchon S, Cao X, Barlowe C, Pelham HR: Got1p and Sft2p: membrane proteins involved in traffic to the Golgi complex. EMBO J. 1999, 18 (14): 3934-3946. 10.1093/emboj/18.14.3934
Valentine MT, Gilbert SP: To step or not to step? How biochemistry and mechanics influence processivity in Kinesin and Eg5. Curr Opin Cell Biol. 2007, 19 (1): 75-81. 10.1016/j.ceb.2006.12.011
Gao S, Alarcon C, Sapkota G, Rahman S, Chen PY, Goerner N, Macias MJ, Erdjument-Bromage H, Tempst P, Massague J: Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-beta signaling. Mol Cell. 2009, 36 (3): 457-468. 10.1016/j.molcel.2009.09.043
Giubettini M, Asteriti IA, Scrofani J, De Luca M, Lindon C, Lavia P, Guarguaglini G: Control of Aurora-A stability through interaction with TPX2. J Cell Sci. 2011, 124 (Pt 1): 113-122.
Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD: WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005, 121 (6): 859-872. 10.1016/j.cell.2005.03.036
Addepalli MK, Ray KB, Kumar B, Ramnath RL, Chile S, Rao H: RNAi-mediated knockdown of AURKB and EGFR shows enhanced therapeutic efficacy in prostate tumor regression. Gene Ther. 2010, 17 (3): 352-359. 10.1038/gt.2009.155
Li Y, Zhu Z, Zhang S, Yu D, Yu H, Liu L, Cao X, Wang L, Gao H, Zhu M: ShRNA-targeted centromere protein A inhibits hepatocellular carcinoma growth. PLoS One. 2011, 6 (3): e17794- 10.1371/journal.pone.0017794
Liu M, Yu H, Huo L, Liu J, Li M, Zhou J: Validating the mitotic kinesin Eg5 as a therapeutic target in pancreatic cancer cells and tumor xenografts using a specific inhibitor. Biochem Pharmacol. 2008, 76 (2): 169-178. 10.1016/j.bcp.2008.04.018
Li B, Qi XQ, Chen X, Huang X, Liu GY, Chen HR, Huang CG, Luo C, Lu YC: Expression of targeting protein for Xenopus kinesin-like protein 2 is associated with progression of human malignant astrocytoma. Brain Res. 2010, 1352: 200-207.
Mattioni T, Hume CR, Konigorski S, Hayes P, Osterweil Z, Lee JS: A cDNA clone for a novel nuclear protein with DNA binding activity. Chromosoma. 1992, 101 (10): 618-624. 10.1007/BF00360539
Sun CT, Lo WY, Wang IH, Lo YH, Shiou SR, Lai CK, Ting LP: Transcription repression of human hepatitis B virus genes by negative regulatory element-binding protein/SON. J Biol Chem. 2001, 276 (26): 24059-24067. 10.1074/jbc.M101330200
Ahn EY, Yan M, Malakhova OA, Lo MC, Boyapati A, Ommen HB, Hines R, Hokland P, Zhang DE: Disruption of the NHR4 domain structure in AML1-ETO abrogates SON binding and promotes leukemogenesis. Proc Natl Acad Sci U S A. 2008, 105 (44): 17103-17108. 10.1073/pnas.0802696105
Sharma A, Takata H, Shibahara K, Bubulya A, Bubulya PA: Son is essential for nuclear speckle organization and cell cycle progression. Mol Biol Cell. 2010, 21 (4): 650-663. 10.1091/mbc.E09-02-0126
Ahn EY, DeKelver RC, Lo MC, Nguyen TA, Matsuura S, Boyapati A, Pandit S, Fu XD, Zhang DE: SON controls cell-cycle progression by coordinated regulation of RNA splicing. Mol Cell. 2011, 42 (2): 185-198. 10.1016/j.molcel.2011.03.014
Leser GP, Fakan S, Martin TE: Ultrastructural distribution of ribonucleoprotein complexes during mitosis. snRNP antigens are contained in mitotic granule clusters. Eur J Cell Biol. 1989, 50 (2): 376-389.
Prasanth KV, Sacco-Bubulya PA, Prasanth SG, Spector DL: Sequential entry of components of the gene expression machinery into daughter nuclei. Mol Biol Cell. 2003, 14 (3): 1043-1057. 10.1091/mbc.E02-10-0669
Bubulya PA, Prasanth KV, Deerinck TJ, Gerlich D, Beaudouin J, Ellisman MH, Ellenberg J, Spector DL: Hypophosphorylated SR splicing factors transiently localize around active nucleolar organizing regions in telophase daughter nuclei. J Cell Biol. 2004, 167 (1): 51-63. 10.1083/jcb.200404120
Lamond AI, Spector DL: Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003, 4 (8): 605-612. 10.1038/nrm1172
Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A: Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010, 464 (7291): 1067-1070. 10.1038/nature08956
Kuboki Y, Shiratori K, Hatori T, Fujita I, Kimijima A, Yamamoto M, Kobayashi M, Furukawa T: Association of epidermal growth factor receptor and mitogen-activated protein kinase with cystic neoplasms of the pancreas. Mod Pathol. 2010, 23 (8): 1127-1135. 10.1038/modpathol.2010.97
Acknowledgements
We thank Dr. Ming S. Tsao at Princess Margaret Hospital and Ontario Cancer Institute, Toronto, ON, for providing HPDE. This study was supported by Grant-in-Aid for Scientific Research (B), Japan Society for the Promotion of Science; Program for Promoting the Establishment of Strategic Research Centers, Special Coordination Funds for Promoting Science and Technology, Ministry of Education, Culture, Sports, Science and Technology (MEXT); and MEXT-Supported Program for the Strategic Research Foundation at Private Universities.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
TF applied a patent on siRNAs used in this study. Other authors declare that they have no competing interests.
Authors’ contribution
TF designed the study. TF and ET carried out in vitro and in vivo experiments and analyzed data. TF, YK, TH, MY, KShim, NS and KShir obtained, examined and analyzed surgical materials. TF wrote the manuscript. All authors had final approval of the submitted and published versions.
Electronic supplementary material
12943_2012_1079_MOESM1_ESM.pdf
Additional file 1: Table S1. Short interfering RNAs used in a systematic knockdown screening of MAPK-assoicated genes in pancreatic cancer. (PDF 37 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Furukawa, T., Tanji, E., Kuboki, Y. et al. Targeting of MAPK-associated molecules identifies SON as a prime target to attenuate the proliferation and tumorigenicity of pancreatic cancer cells. Mol Cancer 11, 88 (2012). https://doi.org/10.1186/1476-4598-11-88
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1476-4598-11-88