Abstract
Background
Anopheles subpictus sensu lato, a widespread malaria vector in Asia, is reportedly composed of four sibling species A - D. Mosquitoes morphologically identified as belonging to the Subpictus complex were collected from different locations near the east coast of Sri Lanka, and specific ribosomal DNA sequences determined to validate their taxonomic status.
Methods
Anopheles subpictus s.l. larvae and blood-fed adults were collected from different locations in the Eastern province and their sibling species status was determined based on published morphological characteristics. DNA sequences of the D3 domain of 28 S ribosomal DNA (rDNA) and the internal transcribed spacer -2 (ITS-2) of mosquitoes morphologically identified as An. subpictus sibling species A, B, C and D were determined.
Results
Phylogenetic analysis based on D3 domain of rDNA resulted in two clades: one clade with mosquitoes identified as An. subpictus species A, C, D and some mosquitoes identified as species B, and another clade with a majority of mosquitoes identified as species B with D3 sequences that were identical to Anopheles sundaicus cytotype D. Analysis of ITS-2 sequences confirmed a close relationship between a majority of mosquitoes identified as An. subpictus B with members of the An. sundaicus complex and others identified as An. subpictus B with An. subpictus s.l.
Conclusions
The study suggests that published morphological characteristics are not specific enough to identify some members of the Subpictus complex, particularly species B. The sequences of the ITS-2 and D3 domain of rDNA suggest that a majority that were identified morphologically as An. subpictus species B in the east coast of Sri Lanka, and some identified elsewhere in SE Asia as An. subpictus s.l., are in fact members of the Sundaicus complex based on genetic similarity to An. sundaicus s.l. In view of the well-known ability of An. sundaicus s.l. to breed in brackish and fresh water and its proven ability to transmit malaria in coastal areas of many Southeast Asian countries, the present findings have significant implications for malaria control in Sri Lanka and neighbouring countries.
Similar content being viewed by others
Background
Anopheles culicifacies species E is the major vector of malaria in Sri Lanka [1]. However Anopheles subpictus s.l. also plays a role in transmitting Plasmodium vivax and Plasmodium falciparum in many parts of Sri Lanka [1–4] as well as in Southeast Asia and India [5, 6]. The taxon An. subpictus is reported to be a species complex comprising four members viz. species A, B, C and D in India [5, 7]. The four members of this complex have been described as possessing characteristic paracentric fixed inversions on the X-chromosome viz. species A (X+a, +b), species B (Xa, b), species C (Xa, +b) and species D (X+a, b) [5, 7]. Additionally, stage-specific morphometric characteristics e.g. the number of ridges in egg floats, larval mesothoracic seta 4, pupal setae and ornamentation of the palpi of adult females, have been reported to be useful for differentiating An. subpictus sibling species in field studies [7]. Initial studies based on a single inversion in the X chromosome suggested the presence of An. subpictus species A and B in Sri Lanka [8]. Subsequent studies based on the numbers of egg ridges indicated the presence of all four sibling species in the country [9]. Members of the complex differ in breeding site and feeding preference and seasonal abundance. The bio-ecological differences between members of the An. subpictus complex in Sri Lanka [1] and elsewhere [5] have been recently reviewed.
Although all four species of An. subpictus are reported to breed in fresh and brackish waters in India, species B is mainly reported to breed in brackish waters [5, 10] and predominate in coastal areas [7, 11]. The coastal populations of the Subpictus complex have been particularly incriminated as malaria vectors in India [12, 13] and Sri Lanka [8]. An. subpictus, including species B, has previously been detected in Eastern Sri Lanka [14, 15]. An. subpictus species B has been specifically implicated in transmitting malaria in the coastal areas of the Puttalam district in the west coast of Sri Lanka [8].
The predominantly brackish water breeding Anopheles sundaicus s.l. is a major vector of malaria in coastal areas of Southeast Asia and Bangladesh [16, 17]. Anopheles sundaicus s.l. was also an important vector of malaria along the east coast of India, including the state of Tamil Nadu (situated in close proximity to Sri Lanka) until DDT spraying largely eliminated it from mainland India, confining it more to the Andaman and Nicobar islands [17, 18]. Anopheles sundaicus s.l. has never previously been detected in Sri Lanka. The taxon An. sundaicus is reported to exist as a species complex comprising four cytological forms (A-D) and four molecular forms namely An. sundaicus s.s., Anopheles epiroticus, An. sundaicus cytotype D and An. sundaicus E [17–19] that occupy fresh and brackish water habitats.
Anopheles vagus is a species related to An. subpictus s.l. and An. sundaicus s.l. that is classified under the same subgenus Cellia and series Pyretophorous. It has been implicated as a secondary malaria vector elsewhere in the Indian subcontinent [20] and is present in Sri Lanka [3, 21].
Morphological properties are widely used to identify and classify mosquito species in field studies because large numbers of samples can be processed without the need for specialized and expensive equipment, which is a common drawback in resource-limited laboratories. However genetic characterization provides the more definitive identification. DNA sequences that give rise to ribosomal RNA (rRNA) are widely used for phylogenetic analysis of closely related organisms including mosquitoes [22, 23]. This is because eukaryotic organisms contain many tandemly repeated copies of rRNA genes that tend to homogenize nucleotide sequences within panmictic species but diverge in sequence between reproductively isolated species. In this context, the expansion loops of 28 S ribosomal RNA, such as the D3 domain, show significantly greater variability between species than core structures. Even greater variability due to more rapid evolution is found in the non-coding internally transcribed spacer (ITS) sequences such as ITS-2, which lies between the 5.8 S and 28 S RNA genes. The D3 and ITS-2 sequences of mosquitoes morphologically identified as An. subpictus species B collected from different coastal locations in the Eastern province of Sri Lanka, and the importance of the findings for understanding malaria transmission in Sri Lanka and elsewhere in Asia are reported here.
Methods
Mosquito collection and identification of sibling species of An. subpictus
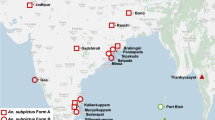
Blood-fed adult anopheline mosquitoes were collected in January and August 2009 from four collection sites in three locations viz. Oluvil (one site within ≈2 km from the coast; 7° 21" 44.41' N: 81° 50" 42.68' E), Chenkalady (one site ≈ 4 km from the coast; 7° 33" 54.93' N: 81° 36" 44.61' E) and Muthur (two sites: an inland area ≈ 8 km from the coast: 8° 25" 29.48' N: 81° 16" 00.97' E; and another < 1 km from the coast: 8° 27" 23.90' N: 81° 15" 33.91' E) in the respective districts of Ampara, Batticaloa and Trincomalee of the Eastern province of Sri Lanka (Figure 1) using cattle baited hut and cattle baited net collections. Larval forms were also collected from a brackish water body in Kallady (< 1 km from the coast and contiguous with the Batticaloa lagoon; 7°43'07.16" N; 81°42'31.28" E) (Figure 1) as previously described [14].
The collected adults were brought to the Zoology laboratory of the Eastern University and identified as An. subpictus s.l. using published keys [6, 24]. Prominent characteristics used for identification were wings with contrasting pale and dark scales, leading margin of wing with at least 4 separate dark areas characteristic of Subgenus Cellia, hind tarsomers 3, 4 and 5 not entirely white scaled, femur and tibia not speckled, foretarsi with broad pale bands crossing joints, palps with preapical dark band equal to or slightly less than the length of apical pale band. Larvae collected at Kallady were screened for reported characteristics of mesothoracic seta 4 (seta 4 M; bifurcate for An. subpictus species A and B but the latter with shorter and thinner branches) for identifying sibling species of the Subpictus complex [7], and reared in the laboratory until they became adults. Facilities and expertise were not available for determining polytene chromosome banding patterns during this investigation and therefore only morphological characteristics were used in identification. Identified blood-fed females were maintained individually and single female F1 progenies were raised as described previously [25]. Sibling species status of females laying eggs was determined through the reported number of ridges in the floats of egg that characterize the different species i.e. species A, 31-36; species B, 16-20; species C, 25-29 and species D, 21-24 [7]. Five to ten eggs from each female were placed on a clean microscopic slide and the number of ridges on floats was counted under a light microscope (x4, Olympus). Adult emerging from identified isofemale progenies were used to examine the ornamentation of wing and palpi. Adult F1 progenies of identified sibling species A, B, C and D were coded and preserved in microcentrifuge tubes containing silica gel plugged with a small piece of cotton wool for molecular studies. Similarly adults that emerged from collected larvae were also preserved separately for molecular characterization.
PCR amplification, DNA sequencing and phylogenetic analysis
The DNA of individual mosquitoes was isolated following the method described by Livak [26]. The ITS-2 region of the rDNA was amplified using primers ITS-2A (5' - TGT GAA CTG CAG GAC ACA T- 3') and ITS-2B (5'- TAT GCT TAA ATT CAG GGG GT - 3') [27]. The D3 domain of 28 S rDNA was amplified using D3A (5'-GAC CCG TCT TGA AAC ACG GA-3') and D3B (5'- TCG GAA GGA ACC AGT TAC TA - 3') primers [28]. For each PCR assay, 50 μl PCR reaction mixture was used. Each mixture contained 0.50 μM of each primer, 200 μM of each dNTP, 1.5 mM of MgCl2 and 1.25 unit of taq polymerase (AmpliTaq Gold, Applied Biosystem). The PCR conditions for both PCRs were an initial denaturation at 95°C for 5 min followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s followed by a final extension at 72°C for 7 min. The PCR products that were successfully amplified were then purified using the Quiaquik PCR Purification Kit (Qiagen, California, USA) to remove unincorporated primers and dNTPs prior to sequencing. The purified PCR amplicons were sent to M/s Macrogen Inc., South Korea, for sequencing in both the forward and reverse directions.
The derived sequences obtained were then analysed together with other representative sequences obtained from GenBank using ClustalW. Neighbour-Joining phylogeny tree was constructed with Bootstrap values from 500 replicates using the MEGA software [29]. Details of the collection sites of different Sri Lankan An. subpictus specimens from the present study and that of An. vagus and other members of the Subpictus and Sundaicus complexes obtained from GenBank and used for phylogenetic analysis of DNA sequences are also summarized in Table 1.
Results
Identification of An. subpictus sibling species and morphological variations in sibling species B
A total of 246 blood-fed An. subpictus s.l. were collected from the four field sites in this study and based on number of ridges on the laid eggs, 2, 152, 11 and 6 females were tentatively characterized as belonging to species A, B, C and D respectively. Different combinations of variant patterns in wing ornamentation (variation in the costal pre-humeral dark spot and pre-apical dark spot) and palpal ornamentation (relative lengths of the apical pale and sub-apical dark bands) in those mosquitoes identified as An. subpictus species B were observed. In the larval collection from Kallady, all 54 adults that emerged from collected larvae were tentatively identified as species B, based on morphological characteristics of larvae and adults.
Phylogenetic analysis based on D3 rRNA
Construction of a phylogenetic tree based on D3 rDNA sequences (Additional file 1) resulted in three stable clades (bootstrap values ≥ 95%). The D3 rDNA phylogenetic tree analysis is presented in Additional file 2. One clade consisted of 14 mosquitoes identified as An. subpictus species A, C, or D and seven identified as species B collected in the present study (LK-B1, 4, 8, 12, 27, 29 and 30) and is termed the An. subpictus clade. All members in this clade possessed identical D3 sequences.
A second clade, termed the An. sundaicus clade, contained 22 mosquitoes identified as An. subpictus species B (LK- B2, 3, 5, 7, 9-11, 13-26, 28) based on the same morphological characteristics as the species B members falling into the An. subpictus clade. The D3 rDNA sequences of all members identified as An. subpictus species B in the An. sundaicus clade (Additional file 2) were identical to those of An. sundaicus cytotype D samples (AY691512, AY691513 and AY691516) collected from the Andaman and Nicobar islands in an unrelated study [30]. The differences between the An. subpictus and An. sundaicus clades were caused by six single nucleotide and two trinucleotide changes and one insertion/deletion within the analysed sequences.
An An. vagus voucher specimen from China (FJ457630.1) formed a third clade that was more closely related to the An. subpictus clade than to the An. sundaicus clade (Additional file 2).
Phylogenetic analysis based on ITS-2 rRNA
Sequence analysis of the ITS-2 sequences of the presumed An. subpictus species B collected in the Eastern province in this study and other available GenBank sequences for An. subpictus s.l. and An. sundaicus s.l. (Additional file 3) were also carried out. The results showed that all species B-type specimens from this study (LK-B series) that formed the An. sundaicus clade with members of Sundaicus complex based on their D3 sequence (LK-B7, 9-11, 16, and 22-24) also stably clustered with An. sundaicus s. l. (AF 369562 and AF 662258) and An. epiroticus based on ITS-2 sequences (Figure 2). This clade is termed the An. sundaicus clade. The only two additional An subpictus B-like specimens in this study whose ITS-2 sequences were determined (LK-B6 and 31) were also found to fall into this An. sundaicus clade. Several specimens identified as An. subpictus s.l. from many other locations in Southeast Asia also grouped with the An. sundaicus clade on analysis of the ITS-2 sequences. These included one specimen (AF406615.1) isolated from an unspecified inland site in Sri Lanka and two others collected from unspecified coastal areas of Sri Lanka (AY049004.1 and AF406616.2). The An. sundaicus clade also included specimens classified as An. subpictus s.l. from Myanmar, Vietnam, Cambodia, Indonesia and Thailand. The ITS-2 sequences were identical in all of the An. subpictus species B specimens collected in this study, the LK-B series, from different sites in Eastern Sri Lanka (Additional file 3). These were also identical to the ITS-2 sequence in An. subpictus s.l. from Myanmar (GQ870334.1), differed from An. subpictus s.l (AF406615.1) in the absence of a single insertion present in the latter, and differed from the two other independent An. subpictus s.l. isolates from Sri Lanka (AY049004.1 and AF406616.2) in three nucleotide changes that were common to the latter (Additional file 3).
Phylogenetic analysis based on ITS-2 of the rDNA. LK- B series refer to individual specimens collected in the study that were initially morphologically characterized as belonging to An. subpictus species B. Other sequences of An. sundaicus s.l. (Sund), An subpictus (Subp) and An. vagus (Vagus) from different locations in countries indicated by their standard two letter abbreviations, were obtained from GenBank.
A second ITS-2 clade in this analysis, termed the An. subpictus clade, was composed of inland populations of An. subpictus s.l. from Punjab, India and Sri Lanka isolated by independent researchers and whose sibling status had not been determined. These could conceivably have all belonged to species A, C or D of Subpictus complex. The ITS-2 sequences from specimens identified as An. subpictus species A, C or D that formed the An. subpictus complex on analysis of the D3 sequences were not available. However ITS-2 sequences from two different specimens (LK-A21 and LK-A22) identified as An. subpictus species A in the present collection were identical to those from two An. subpictus s.l. specimens collected from Southern Sri Lanka by others (GQ870337.1 and GQ870336.1) and that clearly fell into the An. subpictus ITS-2 clade on phylogenetic analysis (Figure 2 and Additional file 3).
Two of the available ITS-2 sequences from An. vagus (FJ457631.1, a voucher specimen from Hainan island in China and FJ654649 from Indonesia) formed a separate clade that was more closely related to the An. subpictus clade than the An. sundaicus clade.
While the three major clades were firmly established with bootstrap values ≥ 99%, some of the internal branches within the An. subpictus and An. sundaicus clades had bootstrap values < 70% indicating a lower degree of robustness in the proposed separation (Figure 2).
Discussion
The Subpictus complex, the Sundaicus complex, An. vagus and the recently described An. pseudosundaicus[31] are closely related taxons. They have been reported to differ in the ornamentation of wings and palpi, properties that are commonly used in identification keys of Anopheline mosquitoes [31]. However only the taxons An. subpictus and An. sundaicus are known to exist as species complexes. Morphological characters that are commonly used to distinguish the four taxons are summarized in Additional file 4. A prominent morphological character that differentiates adult An. sundaicus s.l. from An. subpictus s.l. and An. vagus is reported to be the speckling of the femur and tibia of the legs of An. sundaicus s.l. [6, 31]. Specimens identified as An. subpictus species B in the present study did not show prominent speckling in their femur and tibia. There is an overlap in the reported number of egg ridges that distinguish An. subpictus species B and An. sundaicus s.l. [6]. In An. subpictus species B the number of egg ridges vary from 16-20 while it is reported to be about 20 for An. sundaicus s.l. [6]. All four cytotypes (A - D) of the Sundaicus complex can be differentiated by examining the distribution of heterochromatin blocks [32, 33]. On the other hand, X- chromosomal inversion polymorphism and egg morphology are generally used to discriminate members of the Subpictus complex [5]. The existence of adult (length variation of apical pale band and subapical dark band of female palpi), larval (characteristic of seta 4 M) and pupal (setae of 6, 7, 9-I) morphological variations among members of the Subpictus complex have been reported (Additional file 4) [7, 34].
Importantly, genetic analysis of specimens morphologically identified as An. subpictus B in this study showed that a majority are probably members of the Sundaicus complex distinct from species A, C and D of the An. subpictus complex as well as An. vagus. This group of An. sundaicus s.l. - like mosquitoes were probably incorrectly identified as An. subpictus B based on morphological characteristics. A minority of specimens identified as An. subpictus species B in the present study based on the number of ridges in egg floats or ornamentation, and which were genetically related based on D3 sequences to species A, C and D, may actually be species A, C or D of the Subpictus complex. This is because the reported differences between Indian species B and D on the number of egg ridges and between Indian species B, C and D in adult ornamentation are small [7]. The morphological differences between An. subpictus species A - D reported to exist in India may also not be altogether applicable to Sri Lankan An. subpictus species due to morphological changes consequent to isolation in an island. There is an alternative possibility that the specimens that were morphologically like the Indian An. subpictus species B but with D3 sequences related to species A, C and D might belong to the archetypical An. subpictus species B category originally identified in India through differential X-chromosome morphology [5, 7]. Further chromosomal, molecular and morphological studies are needed to clarify these possibilities.
The genetic data is consistent with a panmictic population of An. sundaicus sibling species being present in the Eastern province of Sri Lanka. The identity of the ITS-2 sequence of this species with that from a specimen identified as An. subpictus s.l. from Myanmar suggests that the latter may belong to the same species. This is consistent with the relative coastal proximity and historically close maritime contacts between Eastern Sri Lanka and Myanmar. One other hand, An. subpictus s.l specimen collected previously from elsewhere in Sri Lanka differed from the An. sundaicus LK-B series sequence in a single insertion while two other specimens showed three nucleotide changes. Hence it is possible that there is some reproductive isolation among An. sundaicus s.l. species in Sri Lanka that has led to differences in the ITS-2 sequences. In this context it will be important to examine the D3 and ITS-2 sequences of mosquitoes previously identified as An. subpictus species B in the Puttalam district in the West coast of Sri Lanka where they are reported to be the major vector of coastal malaria [8]. The present findings also suggest that the two genotypes, An. subpictus s.l. and An. sundaicus s.l. may occur sympatrically in habitats close to the coast.
There is also the indication from available ITS-2 data that members of the Sundaicus complex may have been mistakenly identified as An. subpictus s.l. in other parts of Southeast Asia. Compatible conclusions were reached with a cryptic species tentatively termed An. vagus genotype B in Timor Leste based on morphological characterization [35]. The An. vagus genotype B, was morphologically similar in the absence of speckling in its femur and tibia to An. vagus and An. subpictus s.l. than to An. sundaicus s.l. However it is clustered with An. sundaicus s.l. as well as some specimens identified morphologically as An. subpictus s.l. populations of Sri Lanka on phylogenetic analysis of its ITS-2 sequence [35]. Indeed it is possible that most, if not all, of the reported An. subpictus species B like-mosquitoes in Sri Lanka, and perhaps elsewhere in Southeast Asia, are in fact members of the An. sundaicus complex. Detailed studies of chromosome inversions and molecular genotyping of Asian An. subpictus species B-like mosquitoes is needed to establish whether this is indeed the case.
Morphological characterization of mosquitoes provides a simple, inexpensive and readily applicable way of identifying many malaria vectors in the field. However, there are clearly limitations with using morphological characteristics alone to differentiate anopheline mosquitoes where they exist sympatically as species complexes or as closely related species. In the context of malaria transmission, further molecular characterization utilizing additional mitochondrial genes such as cytochrome oxidase subunits (CO) and cytochrome B (Cyt-b) are additionally needed to characterize the presence of sibling species in Subpictus complex and Sundaicus complex in Asia. This will aid the development of more robust morphology-based or DNA-based identification methods that would be suitable for field-based applications.
Conclusions
Sequence analysis of ITS-2 and 28 S RNA D3 domains of rDNA show that a majority of specimens morphologically identified as An. subpictus species B in Eastern Sri Lanka are in fact members of the Sundaicus complex. The existing morphological variations and their use for identifying closely related anopheline mosquitoes, especially when they exist as species complexes, are imprecise and need to be replaced with DNA sequence-based techniques. The results also suggest that molecular techniques for classifying the relationships between members of the Sundaicus and Subpictus complexes are particularly important in South and Southeast Asia where members of the Sundaicus complex may have a hitherto unappreciated geographical spread helped possibly by long-existing maritime links between the nations of the region. Members of the Sundaicus complex, that are well known malaria vectors in coastal areas, pose the risk of increasing malaria transmission with a rise in sea levels due to global warming and expanding aquaculture. These findings have a direct bearing on the continued development of effective malaria vector control or elimination programmes in Sri Lanka and elsewhere in Asia.
References
Surendran SN, Ramasamy R: The Anopheles culicifacies and An. subpictus complexes in Sri Lanka and their implications for malaria control in the country. Trop Med Health. 2010, 38: 1-11. 10.2149/tmh.2009-12.
Amerasinghe PH, Amerasinghe FP, Wirtz RA, Indrajith NG, Somapala W, Preira LR, Rathnayake AMC: Malaria transmission by Anopheles subpictus Grassi in a new irrigation project in Sri Lanka. J Med Entomol. 1992, 29: 577-581.
Ramasamy R, Ramasamy MS, Wijesundera DA, Wijesundera APdeS, Dewit I, Ranasinghe C, Srikrishnarajah KA, Wickramarantne C: High seasonal malaria transmission rates in the intermediate rainfall zone of Sri Lanka. Ann Trop Med Parasitol. 1992, 86: 591-600.
Amerasinghe PH, Amerasinghe FP, Kondradsen F, Fonseka KT, Wirtz RA: Malaria vectors in a traditional dry zone village in Sri Lanka. Am J Trop Med Hyg. 1999, 60: 421-429.
Chandra G, Bhattacharjee I, Chatterjee S: A review on Anopheles subpictus Grassi - A biological vector. Acta Trop. 2010, 115 (1-2): 142-54. 10.1016/j.actatropica.2010.02.005. Epub 2010 Feb 11
Christophers SR: The Fauna of British India, including Ceylon and Burma. Diptera 4. Family Culicidae. Tribe Anopheline. 1933, Taylor & Francis, London
Suguna SG, Rathinam KG, Rajavel AR, Dhanda V: Morphological and chromosomal descriptions of new species in the Anopheles subpictus complex. Med Vet Entomol. 1994, 8: 88-94. 10.1111/j.1365-2915.1994.tb00392.x.
Abhayawardana TA, Wijesuria SRE, Dilrukshi RRKC: Anopheles subpictus complex: distribution of sibling species in Sri Lanka. Indian J Malariol. 1996, 33: 53-60.
Abhayawardana TA, Wickramasinghe MB, Amerasinghe FP: Sibling species of Anopheles subpictus and their seasonal abundance in Chilaw area. Proc Sri Lanka Assoc Advan Sci. 1999, 55: 17-
Singh SP, Raghavendra K, Kumar R, Nanda N, Subbarao SK: Moprhotaxonomic studies to identify the members of the Anopheles subpictus Grassi (Diptera: Culicidae) species complex in reverine village of district of Sonepat, Haryana state, India. J Commun Dis. 2004, 36: 35-40.
Reuban R, Kalyanasundaram M, Suguna G: Salinity tolerance of sibling species in the taxon Anopheles subpictus Grassi, 1899. Indian J Med Res. 1984, 80: 67-70.
Panicker KN, Geetha BM, Bheema RUS, Wiswam K, Suryanarayana MU: Anopheles subpictus: vector of malaria in costal villages of southeast India. Curr Sci. 1981, 50: 694-5.
Sahu SS: Comparative susceptibility of Anopheles subpictus from fresh and brackish water areas to Plasmodium falciparum infection. Acta Trop. 1998, 70: 1-7. 10.1016/S0001-706X(97)00140-X.
Jude PJ, Dharshini S, Vinobaba M, Surendran SN, Ramasamy R: Anopheles culicifacies breeding in brackish waters in Sri Lanka and implications for malaria control. Malar J. 2010, 9: 106-10.1186/1475-2875-9-106.
Sujarahini K, Ramasamy MS, Dharmaratnam M: Night human biting mosquitoes during the northeast monsoon in Batticaloa (dry zone) of Sri Lanka. Jsc-EUSL. 2001, 2: 16-25.
Dusfour I, Blondeau J, Vythilingam I, Baimai V, Trung H, Sochanta T, Bangs MJ, Manguin S: Polymerase chain reaction identification of three members of the Anopheles sundaicus (Diptera: Culicidae) complex, malaria vectors in Southeast Asia. J Med Entomol. 2007, 44: 723-731. 10.1603/0022-2585(2007)44[723:PCRIOT]2.0.CO;2.
Manguin S, Garros C, Dusfour I, Harbach RE, Coosemans M: Bionomics, taxonomy, and distribution of the major vector taxa of Anopheles subgenus Cellia in Southeast Asia: An updated review. Infect Genet Evol. 2008, 8: 489-503. 10.1016/j.meegid.2007.11.004.
Das MK, Wattal S, Nanda N, Adak T: Laboratory colonization of Anopheles sundaicus. Curr Sci. 2004, 86: 1069-1070.
Bora H, Das MK, Ahmed A, Sharma YD: Variations in the mitochondrial DNA markers in the Anopheles (Cellia) sundaicus population from the Andaman and Nicobar Islands, India. Acta Trop. 2009, 112: 120-124. 10.1016/j.actatropica.2009.07.007.
Alam MS, Khan MGM, Chaudury N, Deloer S, Nazib F, Bangali A, Haque R: Prevalence of anopheline species and their Plasmodium infections status in epidemic-prone border areas of Bangladesh. Malar J. 2010, 9: 15-10.1186/1475-2875-9-15.
Ramasamy R, de Alwis R, Wijesundere A, Ramasamy MS: Malaria transmission at a new irrigation project in Sri Lanka: the emergence of Anopheles annularis as a major vector. Am J Trop Med Hyg. 1992, 47: 547-553.
Singh OP, Nanda N, Dev V, Bali P, Sohali M, Mehrunnisa A, Adak T, Dash AP: Molecular evidence of misidentification of Anopheles minimus as Anopheles fluviatilis in Assam (India). Acta Trop. 2010, 113: 241-244. 10.1016/j.actatropica.2009.11.002.
Collins FH, Paskewitz SM: A review of the use of ribosomal DNA (rDNA) to differentiate among cryptic Anopheles species. Insect Mol Boil. 1996, 5: 1-9. 10.1111/j.1365-2583.1996.tb00034.x.
Amerasinghe FP: A guide to the identification of the anopheline mosquitoes (Diptera: Culicidae) of Sri Lanka -I Adult females. Ceylon J Sci (Bio Sci). 1990, 21: 1-16.
Surendran SN, Ramasamy MS, De Silva BGDNK, Ramasamy R: Anopheles culicifacies sibling species B and E in Sri Lanka differ in longevity and in their susceptibility to malaria parasite infection and common insecticides. Med Vet Entomol. 2006, 20: 153-156. 10.1111/j.1365-2915.2006.00601.x.
Livak KJ: Organization and mapping of a sequence on the Drosophila melanogaster X and Y chromosomes that is transcribed during spermatogenesis. Genetics. 1984, 107: 611-634.
Beebe NW, Saul A: Discrimination of all members of the Anopheles punctulatus complex by polymerase chain reaction - restriction fragment length polymorphism analysis. Am J Trop Med Hyg. 1995, 53 (5): 478-481.
Litvaitis MK, Nunn G, Thomas WK, Kocher TD: A molecular approach for the identification of Meiofaunal turbellarians (Platyhelminthes, Turbellaria). Mar Biol. 1994, 120: 437-442. 10.1007/BF00680218.
Tamara K, Dudley J, Nei M, Kumar S: MEGA4: Molecular evolutionary genetic analysis (MEGA) software version 4.0. Mol Biol Evol. 2007, 24: 1596-1599. 10.1093/molbev/msm092.
Alam MT, Das MK, Ansari MA, Sharma YD: Molecular identification of Anopheles (Cellia) sundaicus from the Andaman and Nicobar islands of India. Acta Trop. 2006, 97: 10-18. 10.1016/j.actatropica.2005.07.028.
Tyagi BK, Hiriyan J, Tewari SC, Ayanar K, Samuel PP, Arunachalam N, Paramasivan R, Krishnamoorthy R, Dhananjeyan K, Leo SV, Rajendran R: Description of a new species, Anopheles pseudosundaicus (Diptera: Culicidae) from Kerala, India. Zootaxa. 2009, 2219: 49-60.
Sukowati S, Baimai V: A standard cytogenetic map for Anopheles sundaicus (Diptera: Culicidae) and evidence for chromosomal differentiation in populations from Thailand and Indonesia. Genome. 1996, 39: 165-173. 10.1139/g96-022.
Nanda N, Das MK, Wattal S, Adak T, Subbarao SK: Cytogenetic characterization of Anopheles sundaicus (Diptera: Culicidae) population from Car Nicobar Islands, India. Ann Entomol Soc Am. 2004, 97: 171-176. 10.1603/0013-8746(2004)097[0171:CCOASD]2.0.CO;2.
Tyagi BK: A note on variations in the prehumeral and humeral accessory dark spots in the wing-costa of the mosquito Anopheles subpictus Grassi, 1899 from south Gujarat, India. Geobios New Reports. 1985, 4: 56-57.
Cooper RD, Edstein MD, Frances SP, Beebe NW: Malaria vectors of Timor-Leste. Malar J. 2010, 9: 40-10.1186/1475-2875-9-40.
Acknowledgements
The authors thank Mr. N. Pirashanthan (University of Jaffna) for Figure 1 and Dr. M. Vinobaba (Department of Zoology, Eastern University of Sri Lanka) for logistic support. The study was partially supported by the National Science Foundation of Sri Lanka (SIDA/BT/2006/03).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SNS and RR conceived the study. PJJ and SNS performed all the field and laboratory studies. OPS did sequence analysis and RR performed phylogenetic analysis. RR and SNS wrote the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
12936_2010_1425_MOESM1_ESM.DOC
Additional file 1: D3 sequences used for phylogenetic analysis. Phylogenetic analysis is based on the D3 region of 28 S rDNA sequences. LK- A, B, C and D series that refer to individual specimens collected in the present study in Sri Lanka that were initially morphologically characterized as belonging to An. subpictus species A, B, C and D respectively. The sequences of An. sundaicus (Sund) from Andaman and Nicobar islands of India (IN) and An. vagus (Vagus) from China were obtained from GenBank. (DOC 42 KB)
12936_2010_1425_MOESM2_ESM.DOC
Additional file 2: Phylogenetic analysis based on the D3 region of 28 S rDNA sequences. The analysis is based on LK-A, B, C and D series that were individual specimens collected in the study that were initially characterized as belonging to An. subpictus A, B, C and D respectively. The sequences of An. sundaicus (Sund) from Andaman and Nicobar silands of India (IN) and An. vagus (Vagus) from China were obtained from GenBank. (DOC 26 KB)
12936_2010_1425_MOESM3_ESM.DOC
Additional file 3: ITS-2 sequences used for phylogenetic analysis. Sequences of the ITS-2 region of rDNA used for phylogenetic analysis. LK- B series refer to individual specimens collected in the study that were initially morphologically characterized as belonging to An. subpictus species B. LK-A series refers to those similarly characterized as An, subpictus species A. Other sequences of An. sundaicus s.l. (Sund), An subpictus (Subp) and An. vagus (Vagus) from different locations in countries indicated by their standard two letter abbreviations, were obtained from GenBank. (DOC 52 KB)
12936_2010_1425_MOESM4_ESM.DOC
Additional file 4: Morphological differentiation of An. sundaicus , An. vagus , An. pseudosundaicus and An. subpictus. Reported morphological characteristics that differentiate An. sundaicus, An. vagus, An. pseudosundaicus and An. subpictus populations and their distribution among sibling species of the Subpictus Complex (DOC 32 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Surendran, S.N., Singh, O.P., Jude, P.J. et al. Genetic evidence for malaria vectors of the Anopheles sundaicus complex in Sri Lanka with morphological characteristics attributed to Anopheles subpictus species B. Malar J 9, 343 (2010). https://doi.org/10.1186/1475-2875-9-343
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2875-9-343