Abstract
Background
Severe malaria (SM) syndromes caused by Plasmodium falciparum infection result in major morbidity and mortality each year. However, only a fraction of P. falciparum infections develop into SM, implicating host genetic factors as important determinants of disease outcome. Previous studies indicate that tumour necrosis factor (TNF) and lymphotoxin alpha (LTα) may be important for the development of cerebral malaria (CM) and other SM syndromes.
Methods
An extensive analysis was conducted of single nucleotide polymorphisms (SNPs) in the TNF, LTA and LTB genes in highland Papuan children and adults, a population historically unexposed to malaria that has migrated to a malaria endemic region. Generated P-values for SNPs spanning the LTA/TNF/LTB locus were corrected for multiple testing of all the SNPs and haplotype blocks within the region tested through 10,000 permutations. A global P-value of < 0.05 was considered statistically significant.
Results
No associations between SNPs in the TNF/LTA/LTB locus and susceptibility to SM in highland Papuan children and adults were found.
Conclusions
These results support the notion that unique selective pressure on the TNF/LTA/LTB locus in different populations has influenced the contribution of the gene products from this region to SM susceptibility.
Similar content being viewed by others
Background
Severe malaria (SM) caused by Plasmodium falciparum results in more than a million deaths each year [1, 2]. It is a collection of syndromes that includes cerebral malaria (CM), severe malaria anaemia (SMA), acute respiratory distress syndrome (ARDS), hyperparasitaemia, hypoglycaemia, black water fever, metabolic acidosis, jaundice and renal failure [1]. The reasons why some individuals develop severe complications of malaria, whereas others do not, are still unclear. However, the virulence of the parasite strain causing malaria, as well as the age and genetic background of the infected individual are likely to influence disease outcome.
Plasmodium falciparum and humans have had intense interactions for at least 6,000 years and it is thought that malaria has imposed a large selective pressure on the human genome [3, 4]. Evidence from both an experimental cerebral malaria (ECM) model [5], as well as from SM patients [6–8], have identified TNF as an important pro-inflammatory cytokine for the control of infection, and also a strong association with the development of pathology. Single nucleotide polymorphisms (SNPs) within the gene encoding TNF (TNF) have been associated with severe outcomes following Plasmodium infection in a number of populations in Africa, Asia and the Pacific region [9–13]. In particular, a SNP in the promoter region of TNF at 308 nucleotide base pairs upstream of the transcription start site (-308/376TNF) has been reported to confer a greater risk of severe neurological sequelae or death due to CM in the Gambia [14]. This variant was found to be a stronger transcriptional activator than the common allele in some in vitro studies [15], but not others [16]. More recent studies examining the MHC class III region, which includes TNF as well as the genes encoding the closely related LTα (LTA) and LTβ (LTB), suggest that positive associations between disease and TNF alleles could in fact be due to real disease alleles in neighbouring genes [17]. One such study observed that when specific SNPs were present in TNF, in combination with specific SNPs in LTA, LTA transcription rather than TNF transcription was modulated by changing the way RNA polymerase specifically bound to the LTA promoter [18]. Importantly, LTα, not TNF, has been shown to be a critical factor in the development of experimental cerebral malaria in C57BL/6 mice [19], identifying LTA along with TNF as a candidate susceptibility gene.
Recently, a genome wide association study involving 3 African populations from The Gambia, Kenya and Malawi, investigated potential associations between SM and 8 SNPs spanning the LTA/TNF locus [20]. An association was found between TNF-238A allele and SM in samples from The Gambia only, and not for any other SNP tested in any of the populations examined [20]. These findings suggested different selective pressures in the LTA/TNF locus in different populations, as well as highlighting the need for more-detailed mapping of polymorphisms across this locus to identify causal SNPs associated with SM susceptibility [20]. Here, an extensive analysis was conducted of 35 SNP's found in the LTA/TNF/LTB gene locus in adults and children of highland Papuan origin, a population not historically exposed to malaria transmission prior to the 1970 s, but who during the study period had been exposed to malaria following migration to the lowland region of Papua.
Methods
Study participants and sample preparation
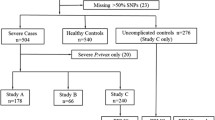
Characteristics of the study participants upon admission to hospital and controls, including population structure, have been described in Table 1 and the methods section of a recent publication [21]. No corrections were required. Highland Papuan patients with SM and asymptomatic malaria-exposed controls were recruited in a case-control study in Timika, a lowland region of Papua, Indonesia. Genotypes significantly associated with SM in highland Papuans, were also examined in a Tanzanian case-control study comprising children with CM enrolled in Dar es Salaam using WHO criteria as previously described [22] and asymptomatic malaria-exposed, healthy control children from Mikocheni Primary school in the Kinondoni Municipality of the Dar es Salaam region. Written informed consent was obtained from all study subjects or their next of kin, parent or guardian. Studies were approved by the Ethics Committees of the National Institute of Health Research and Development (Ministry of Health, Jakarta, Indonesia), Menzies School of Health Research (Darwin, Australia), Queensland Institute of Medical Research (Brisbane, Australia), Muhimbili University of Health Sciences (Dar es Salaam, Tanzania), National Institute for Medical Research (Dar es Salaam, Tanzania), University of Utah Medical Center (Salt Lake City, USA) and Duke University Medical Center (Durham, USA).
Sequencing the LTA gene
The gene encoding LTα, spanning a region of 5323 base pairs (bp) was sequenced in a subset of samples to check for the presence of novel SNPs. The locus was amplified in sections of approximately 500 bp by PCR and sequenced. Sequenced products were aligned against a published human LTA sequence [23]. Identified SNPs were also checked on the NCBI website [24] for previous publication.
SNP selection
TNF/LTA/LTB SNPs were included in this study based on reported functional changes (i.e. transcriptional or protein level) or on previous associations with malaria, other infectious diseases, ischaemic stroke, cerebral infarction, atherosclerosis or inflammation [9–12, 14, 18, 25–43]. SNPs in the TNF/LTA/LTB locus observed to be present in ethnic groups from Sulawesi, an island in the Indonesian archipelago [44], were also included. The final set of polymorphisms included as many functional and disease-associated SNPs as possible following primer design and testing. Tagging SNPs were selected based on reported frequencies in other populations. This SNP set has been described elsewhere [45]. All SNP sequences were obtained from the Chip Bioinformatics database [46] and verified in NCBI.
Genotyping
Assays were designed for thirty-five SNPs across the TNF/LTA/LTB region in a multiplex using the Sequenom MassARRAY Assay Design software (version 3.0). SNPs were typed using iPLEX™ chemistry and analyzed using a Sequenom MassARRAY Compact Mass Spectrometer (Sequenom Inc, San Diego, CA, USA). The 2.5 ml PCR reactions were performed in standard 384-well plates using 10 ng genomic DNA, 0.5 unit of Taq polymerase (HotStarTaq, Qiagen), 500 mmol of each dNTP, and 100 nmol of each PCR primer. Standard PCR thermal cycling conditions and post-PCR extension reactions were carried out as described previously [45]. The iPLEX reaction products were desalted and spotted on a SpectroChip (Sequenom). Data were processed and analysed by MassARRAY Workstation (version 3.4) software (Sequenom). Single SNP genotyping specific for rs2071590 and rs1052248 was performed using TaqMan SNP Genotyping Assays according to the manufacturer's instructions (Applied Biosystems, Foster City, CA) by using the allelic discrimination on a Corbett RG-6000 (Corbett Life Sciences, Sydney, NSW, Australia) or an AB7900 machine (Applied Biosystems). Previously characterized genotypes for these SNPs were included as positive controls and run alongside samples with unknown genotype.
Statistical analyses
Based on our sample size (380 cases and 356 controls), 80% power was available to detect a disease allele with a relative risk of 1.5 at a disease frequency of 0.25. Haploview version 3.32 (Whitehead Institute for Biomedical Research, USA; [47]) was used to perform all statistical tests relating to this SNP analysis [48]. Genotype frequencies of all SNPs were tested for departures from Hardy-Weinberg equilibrium in both cases and controls separately. In the Papuan study, 16 SNPs were found to be non-polymorphic (Table 1). Haplotype frequencies and linkage disequilibrium (LD) tests were also determined by Haploview version 3.32 [48] using the default method of Gabriel [49]. The association between single markers and haplotype blocks was performed by the Haploview programme. Generated P-values for SNPs spanning the LTA/TNF/LTB locus were corrected for multiple testing of all the SNPs and haplotype blocks within the region tested through 10,000 permutations. A global P-value of < 0.05 was considered statistically significant.
Results
LTA/TNF locus is not associated with severe malaria
In total, 380 SM cases (262 adults and 118 children) and 356 control individuals (305 adults and 51 children) were included in the genotyping study [21]. A subset of the samples was randomly chosen for sequencing of a 5,323 bp region spanning LTA, and this revealed no novel SNPs in the study population, and that all SNPs detected in the sequenced LTA had previously been reported and, where possible, were included in the SNP set (Table 1). The SNP set consisted of 35 SNPs that spanned the LTA/TNF/LTB locus on chromosome 6 (Figure 1 and Table 1). The majority of the samples were successfully genotyped for each of the SNPs with an average coverage of greater than 99%. Of these, 16 SNPs were found to be non-polymorphic (Table 1). The minor allele frequencies of the remaining SNPs spanning the LTA/TNF/LTB locus ranged from 0.001 to 0.44 in control subjects and from 0.001 to 0.49 in patients with SM. Two SNPs, rs2071590 and rs1052248, situated in the LTA and LTB promoter regions, respectively, showed some differences in allele frequencies between control subjects and SM patients (P = 0.034 and P = 0.007, respectively). These differences were not observed once corrections were made for multiple testing (global P > 0.05; Table 2). Analysis of haplotypes did not reveal any association between haplotype blocks and susceptibility to SM (Figure 2 and Table 3).
Chromosomal location of SNPs studied in the LTA/TNF/LTB locus. 35 SNPs that cover the LTA/TNF/LTB locus on chromosome 6 were analysed in highland Papuans. SNPs selected for study were based on reported functional changes, previous associations with malaria, and other infectious diseases or inflammatory conditions. Certain tagged SNPs were included based on reported frequencies in other populations.
Linkage disequilibrium plot of LTA/TNF/LTB SNP set in all severe malaria patients. The LD plot shading scheme illustrates the estimated LD between the SNPs, and the LD value is displayed within the box. Dark grey regions signify strong LD (1.0), whereas light grey and white regions depict low LD (< 1.0).
Next, the TNF/LTA/LTB locus was examined to establish whether it was associated with susceptibility to CM in either adults or children. Overall, there were no differences in allele frequencies between control subjects and CM patients (Table 4). Furthermore, neither age nor susceptibility to CM was associated with the TNF/LTA/LTB locus for either single SNPs or haplotype blocks (Tables 3 and 4; Figure 2). Given the association between the two SNPs situated in the LTA and LTB promoter regions and SM prior to correcting for multiple testing described above, the association of rs2071590 and rs1052248 in patients originating from and resident in a malaria-endemic region (245 healthy children (HC) and 77 children with CM from Dar es Salaam, Tanzania) was also examined, but no association between these SNPs and CM was found. Taken together, there was no evidence for the TNF/LTA/LTB locus contributing to SM or CM in highland Papuans historically not exposed to malaria.
Discussion
Both genetic and serological data indicate roles for TNF and LTα in the pathogenesis of SM [6–14, 50, 51]. However, there was a lack of association between SNPs in the TNF/LTA/LTB and SM in a highland Papuan population. This lack of association remained following subsequent testing in disease subsets and in both children and adults. TNF polymorphisms have been associated with malaria transmission and SM, primarily in populations originating and living in malaria endemic areas [9–14]. A small number of studies have investigated the relationship between two LTA SNPs and malaria. LTA C+80A (rs2239704), a SNP that allows specific binding of the transcriptional repressor ABF-1 and, therefore, considered to be a low LTα-producing allele, has been associated with lower P. falciparum parasitaemia in malaria-endemic Burkina Faso but was not associated with SM in a case-control study in The Gambia [40, 51, 52]. LTA A+252G (rs909253) has been reported to influence LTα production [53], but this SNP was not associated with SM in Sri Lanka [9]. More recently, rs2239704 and rs909253 were reported to not be associated with SM in a study of SM patients from The Gambia, Kenya and Malawi [20]. Both rs2239704 and rs909253 were included in the present study but not found to be associated with SM. A recent genome wide association study found that only rs2516486 at TNF was weakly associated with malaria severity, however the authors caution that the candidate SNPs examined in this study were poorly tagged by the 500 K array used [54].
LTA polymorphisms were of particular interest. In highland Papuans (a population without historical exposure to malaria), the minor allele for rs2239704 (Table 3) is the opposite allele to that observed in a study population from malaria-endemic Burkina Faso [51], suggesting that different pressures may have selected for different alleles in the two populations. Despite two SNPs situated in the LTA and LTB promoter regions (rs2071590 and rs1052248, respectively), having differences in allele frequencies between control subjects and SM patients, this was not significant after correction for multiple testing, nor was there a difference in the Tanzanian population studied. Thus, our data provide no evidence for an involvement of the LTA/TNF/LTB locus in SM susceptibility in highland Papuans, and suggest that if the genes encoded by this locus are involved in SM pathogenesis then molecules that regulate the production and/or bioavailablity may influence disease outcome. Interestingly, an association between susceptibility to SM and a SNP in the LTα-related gene encoding galectin-2 (LGALS2) was recently identified. The particular SNP in LGALS2 is thought to regulate the trafficking of LTα out of cells [55]. Strikingly, the association between LGALS2 and SM was found to be present in highland Papuan children, but not adults [21]. A further consideration in future studies should be the effects of other genes with important immunological functions in the region surrounding the TNF locus that could contribute to the development of severe malaria. The potential importance of such genes was highlighted in a recent study showing that SNPs in HLA-B associated transcript 2 (BAT2) in the MHC III region were associated with severe malaria susceptibility, while TNF and LTA SNPs were not [52].
Conclusion
It is clear that large, highly powered studies will be necessary to identify causal variants in genes that contribute to SM disease outcome. However, as discussed above, it is likely that different populations have had different selective pressures placed upon them, resulting in a number of susceptibility alleles across different populations. Hence, deep sequencing studies in the TNF/LTA/LTB locus of different populations may be warranted and required to identify causal variants in these genes responsible for susceptibility to SM syndromes.
Abbreviations
- SM:
-
severe malaria
- TNF:
-
tumour necrosis factor
- LT:
-
lymphotoxin
- CM:
-
cerebral malaria
- SNP:
-
single nucleotide polymorphism
- SMA:
-
severe malaria anaemia
- ARDS:
-
acute respiratory distress syndrome
- bp:
-
base pair
- LD:
-
linkage disequilibrium.
References
WHO: Severe falciparum malaria. World Health Organization, Communicable Diseases Cluster. Trans R Soc Trop Med Hyg. 2000, 94 (Suppl 1): S1-90.
WHO: World Malaria Report 2008. 2008, Program WGM. Geneva
Rich SM, Licht MC, Hudson RR, Ayala FJ: Malaria's Eve: evidence of a recent population bottleneck throughout the world populations of Plasmodium falciparum. Proc Natl Acad Sci USA. 1998, 95: 4425-4430. 10.1073/pnas.95.8.4425.
Kun JF, Mordmuller B, Perkins DJ, May J, Mercereau-Puijalon O, Alpers M, Weinberg JB, Kremsner PG: Nitric oxide synthase 2(Lambarene) (G-954C), increased nitric oxide production, and protection against malaria. J Infect Dis. 2001, 184: 330-336. 10.1086/322037.
Grau GE, Fajardo LF, Piguet PF, Allet B, Lambert PH, Vassalli P: Tumor necrosis factor (cachectin) as an essential mediator in murine cerebral malaria. Science. 1987, 237: 1210-1212. 10.1126/science.3306918.
Grau GE, Piguet PF, Vassalli P, Lambert PH: Tumor-necrosis factor and other cytokines in cerebral malaria: experimental and clinical data. Immunol Rev. 1989, 112: 49-70. 10.1111/j.1600-065X.1989.tb00552.x.
Kern P, Hemmer CJ, Van Damme J, Gruss HJ, Dietrich M: Elevated tumor necrosis factor alpha and interleukin-6 serum levels as markers for complicated Plasmodium falciparum malaria. Am J Med. 1989, 87: 139-143. 10.1016/S0002-9343(89)80688-6.
Kwiatkowski D: Tumour necrosis factor, fever and fatality in falciparum malaria. Immunol Lett. 1990, 25: 213-216. 10.1016/0165-2478(90)90117-9.
Wattavidanage J, Carter R, Perera KL, Munasingha A, Bandara S, McGuinness D, Wickramasinghe AR, Alles HK, Mendis KN, Premawansa S: TNFalpha*2 marks high risk of severe disease during Plasmodium falciparum malaria and other infections in Sri Lankans. Clin Exp Immunol. 1999, 115: 350-355. 10.1046/j.1365-2249.1999.00804.x.
Aidoo M, McElroy PD, Kolczak MS, Terlouw DJ, ter Kuile FO, Nahlen B, Lal AA, Udhayakumar V: Tumor necrosis factor-alpha promoter variant 2 (TNF2) is associated with pre-term delivery, infant mortality, and malaria morbidity in western Kenya: Asembo Bay Cohort Project IX. Genet Epidemiol. 2001, 21: 201-211. 10.1002/gepi.1029.
Ubalee R, Suzuki F, Kikuchi M, Tasanor O, Wattanagoon Y, Ruangweerayut R, Na-Bangchang K, Karbwang J, Kimura A, Itoh K, Kanda T, Hirayama K: Strong association of a tumor necrosis factor-alpha promoter allele with cerebral malaria in Myanmar. Tissue Antigens. 2001, 58: 407-410. 10.1034/j.1399-0039.2001.580610.x.
Hananantachai H, Patarapotikul J, Ohashi J, Naka I, Krudsood S, Looareesuwan S, Tokunaga K: Significant association between TNF-alpha (TNF) promoter allele (-1031C, -863C, and -857C) and cerebral malaria in Thailand. Tissue Antigens. 2007, 69: 277-280. 10.1111/j.1399-0039.2006.00756.x.
Sinha S: TNF-alpha alleles and susceptibility to cerebral malaria. Natl Med J India. 1995, 8: 70-72.
McGuire W, Hill AV, Allsopp CE, Greenwood BM, Kwiatkowski D: Variation in the TNF-alpha promoter region associated with susceptibility to cerebral malaria. Nature. 1994, 371: 508-510. 10.1038/371508a0.
Wilson AG, Symons JA, McDowell TL, McDevitt HO, Duff GW: Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci USA. 1997, 94: 3195-3199. 10.1073/pnas.94.7.3195.
Brinkman BM, Zuijdeest D, Kaijzel EL, Breedveld FC, Verweij CL: Relevance of the tumor necrosis factor alpha (TNF alpha) -308 promoter polymorphism in TNF alpha gene regulation. J Inflamm. 1995, 46: 32-41.
Ackerman HC, Ribas G, Jallow M, Mott R, Neville M, Sisay-Joof F, Pinder M, Campbell RD, Kwiatkowski DP: Complex haplotypic structure of the central MHC region flanking TNF in a West African population. Genes Immun. 2003, 4: 476-486. 10.1038/sj.gene.6364008.
Knight JC, Keating BJ, Rockett KA, Kwiatkowski DP: In vivo characterization of regulatory polymorphisms by allele-specific quantification of RNA polymerase loading. Nat Genet. 2003, 33: 469-475. 10.1038/ng1124.
Engwerda CR, Mynott TL, Sawhney S, DeSouza JB, Bickle QD, Kaye PM: Locally up-regulated lymphotoxin alpha, not systemic tumor necrosis factor alpha, is the principle mediator of murine cerebral malaria. J Exp Med. 2002, 195: 1371-1377. 10.1084/jem.20020128.
Clark TG, Diakite M, Auburn S, Campino S, Fry AE, Green A, Richardson A, Small K, Teo YY, Wilson J, Jallow M, Sisay-Joof F, Pinder M, Griffiths MJ, Peshu M, Williams TN, Marsh K, Molyneux ME, Taylor TE, Rockett KA, Kwiatkowski DP: Tumor necrosis factor and lymphotoxin-alpha polymorphisms and severe malaria in African populations. J Infect Dis. 2009, 199: 569-575. 10.1086/596320.
Randall LM, Kenangalem E, Lampah DA, Tjitra E, Mwaikambo ED, Handojo T, Piera KA, Zhao ZZ, de Labastida Rivera F, Zhou Y, McSweeney KM, Le L, Amante FH, Haque A, Stanley AC, Woodberry T, Salwati E, Granger DL, Hobbs MR, Price RN, Weinberg JB, Montgomery GW, Anstey NM, Engwerda CR: Age-related susceptibility to severe malaria associated with galectin-2 in highland Papuans. J Infect Dis. 2010, 202: 117-124. 10.1086/653125.
Anstey NM, Weinberg JB, Hassanali MY, Mwaikambo ED, Manyenga D, Misukonis MA, Arnelle DR, Hollis D, McDonald MI, Granger DL: Nitric oxide in Tanzanian children with malaria: inverse relationship between malaria severity and nitric oxide production/nitric oxide synthase type 2 expression. J Exp Med. 1996, 184: 557-567. 10.1084/jem.184.2.557.
Ensembl. [http://www.ensembl.org]
National Center for Biotechnology Information. [http://www.ncbi.nlm.nih.gov/SNP]
Um JY, An NH, Kim HM: TNF-alpha and TNF-beta gene polymorphisms in cerebral infarction. J Mol Neurosci. 2003, 21: 167-171. 10.1385/JMN:21:2:167.
Lee BC, Ahn SY, Doo HK, Yim SV, Lee HJ, Jin SY, Hong SJ, Lee SH, Kim SD, Seo JC, Leem KH, Chung JH: Susceptibility for ischemic stroke in Korean population is associated with polymorphisms of the interleukin-1 receptor antagonist and tumor necrosis factor-alpha genes, but not the interleukin-1beta gene. Neurosci Lett. 2004, 357: 33-36. 10.1016/j.neulet.2003.12.041.
Cabrera M, Shaw MA, Sharples C, Williams H, Castes M, Convit J, Blackwell JM: Polymorphism in tumor necrosis factor genes associated with mucocutaneous leishmaniasis. J Exp Med. 1995, 182: 1259-1264. 10.1084/jem.182.5.1259.
Albuquerque RV, Hayden CM, Palmer LJ, Laing IA, Rye PJ, Gibson NA, Burton PR, Goldblatt J, Lesouef PN: Association of polymorphisms within the tumour necrosis factor (TNF) genes and childhood asthma. Clin Exp Allergy. 1998, 28: 578-584. 10.1046/j.1365-2222.1998.00273.x.
Trabetti E, Patuzzo C, Malerba G, Galavotti R, Martinati LC, Boner AL, Pignatti PF: Association of a lymphotoxin alpha gene polymorphism and atopy in Italian families. J Med Genet. 1999, 36: 323-325.
Moffatt MF, James A, Ryan G, Musk AW, Cookson WO: Extended tumour necrosis factor/HLA-DR haplotypes and asthma in an Australian population sample. Thorax. 1999, 54: 757-761. 10.1136/thx.54.9.757.
Grutters JC, Sato H, Pantelidis P, Lagan AL, McGrath DS, Lammers JW, van den Bosch JM, Wells AU, du Bois RM, Welsh KI: Increased frequency of the uncommon tumor necrosis factor -857T allele in British and Dutch patients with sarcoidosis. Am J Respir Crit Care Med. 2002, 165: 1119-1124.
Swider C, Schnittger L, Bogunia-Kubik K, Gerdes J, Flad H, Lange A, Seitzer U: TNF-alpha and HLA-DR genotyping as potential prognostic markers in pulmonary sarcoidosis. Eur Cytokine Netw. 1999, 10: 143-146.
Bouqbis L, Akhayat O, Garchon HJ, Calafell F, Izaabel H: TNFA-TNFB haplotypes modify susceptibility to type I diabetes mellitus independently of HLA class II in a Moroccan population. Tissue Antigens. 2003, 61: 72-79. 10.1034/j.1399-0039.2003.610106.x.
Lio D, Caruso C, Di Stefano R, Colonna Romano G, Ferraro D, Scola L, Crivello A, Licata A, Valenza LM, Candore G, Craxi A, Almasio PL: IL-10 and TNF-alpha polymorphisms and the recovery from HCV infection. Hum Immunol. 2003, 64: 674-680. 10.1016/S0198-8859(03)00080-6.
Niro GA, Fontana R, Gioffreda D, Valvano MR, Lacobellis A, Facciorusso D, Andriulli A: Tumor necrosis factor gene polymorphisms and clearance or progression of hepatitis B virus infection. Liver Int. 2005, 25: 1175-1181. 10.1111/j.1478-3231.2005.01166.x.
Knight JC, Udalova I, Hill AV, Greenwood BM, Peshu N, Marsh K, Kwiatkowski D: A polymorphism that affects OCT-1 binding to the TNF promoter region is associated with severe malaria. Nat Genet. 1999, 22: 145-150. 10.1038/9649.
Ozaki K, Ohnishi Y, Iida A, Sekine A, Yamada R, Tsunoda T, Sato H, Sato H, Hori M, Nakamura Y, Tanaka T: Functional SNPs in the lymphotoxin-alpha gene that are associated with susceptibility to myocardial infarction. Nat Genet. 2002, 32: 650-654. 10.1038/ng1047.
Satoh T, Pandey JP, Okazaki Y, Yasuoka H, Kawakami Y, Ikeda Y, Kuwana M: Single nucleotide polymorphisms of the inflammatory cytokine genes in adults with chronic immune thrombocytopenic purpura. Br J Haematol. 2004, 124: 796-801. 10.1111/j.1365-2141.2004.04843.x.
Suzuki G, Izumi S, Hakoda M, Takahashi N: LTA 252G allele containing haplotype block is associated with high serum C-reactive protein levels. Atherosclerosis. 2004, 176: 91-94. 10.1016/j.atherosclerosis.2003.12.013.
Knight JC, Keating BJ, Kwiatkowski DP: Allele-specific repression of lymphotoxin-alpha by activated B cell factor-1. Nat Genet. 2004, 36: 394-399. 10.1038/ng1331.
Laxton R, Pearce E, Kyriakou T, Ye S: Association of the lymphotoxin-alpha gene Thr26Asn polymorphism with severity of coronary atherosclerosis. Genes Immun. 2005, 6: 539-541. 10.1038/sj.gene.6364236.
Liu Y, Herrington D, Burdon KP, Langefeld CD, Rich SS, Bowden DW, Freedman BI, Wagenknecht LE: A functional polymorphism in the lymphotoxin-alpha gene is associated with carotid artery wall thickness: the Diabetes Heart Study. Eur J Cardiovasc Prev Rehabil. 2006, 13: 655-657. 10.1097/01.hjr.0000214610.83866.2e.
Niwa Y, Hirose K, Matsuo K, Tajima K, Ikoma Y, Nakanishi T, Nawa A, Kuzuya K, Tamakoshi A, Hamajima N: Lymphotoxin-alpha polymorphism and the risk of cervical cancer in Japanese subjects. Cancer Lett. 2005, 218: 63-68. 10.1016/j.canlet.2004.09.021.
Lamsis F, Flannery GR, White NG, Muratore R, Kaelan C, Mitchell RJ: Alleles and haplotypes of tumor necrosis factor (TNF) alpha and beta genes in three ethnic populations of Sulawesi Indonesia. Hum Biol. 2002, 74: 381-396. 10.1353/hub.2002.0030.
Zhao ZZ, Nyholt DR, Le L, Thomas S, Engwerda C, Randall L, Treloar SA, Montgomery GW: Genetic variation in tumour necrosis factor and lymphotoxin is not associated with endometriosis in an Australian sample. Hum Reprod. 2007, 22: 2389-2397. 10.1093/humrep/dem182.
CHIP Bioinformatics Tools. [http://snpper.chip.org]
Haploview. [http://www.broad.mit.edu/mpg/haploview]
Barrett JC, Fry B, Maller J, Daly MJ: Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005, 21: 263-265. 10.1093/bioinformatics/bth457.
Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D: The structure of haplotype blocks in the human genome. Science. 2002, 296: 2225-2229. 10.1126/science.1069424.
Clark IA, Gray KM, Rockett EJ, Cowden WB, Rockett KA, Ferrante A, Aggarwal BB: Increased lymphotoxin in human malarial serum, and the ability of this cytokine to increase plasma interleukin-6 and cause hypoglycaemia in mice: implications for malarial pathology. Trans R Soc Trop Med Hyg. 1992, 86: 602-607. 10.1016/0035-9203(92)90144-2.
Barbier M, Delahaye NF, Fumoux F, Rihet P: Family-based association of a low producing lymphotoxin-alpha allele with reduced Plasmodium falciparum parasitemia. Microbes Infect. 2008, 10: 673-679. 10.1016/j.micinf.2008.03.001.
Diakite M, Clark TG, Auburn S, Campino S, Fry AE, Green A, Morris AP, Richardson A, Jallow M, Sisay-Joof F, Pinder M, Kwiatkowski DP, Rockett KA: A genetic association study in the Gambia using tagging polymorphisms in the major histocompatibility complex class III region implicates a HLA-B associated transcript 2 polymorphism in severe malaria susceptibility. Hum Genet. 2009, 125: 105-109. 10.1007/s00439-008-0597-2.
Messer G, Spengler U, Jung MC, Honold G, Blomer K, Pape GR, Riethmuller G, Weiss EH: Polymorphic structure of the tumor necrosis factor (TNF) locus: an NcoI polymorphism in the first intron of the human TNF-beta gene correlates with a variant amino acid in position 26 and a reduced level of TNF-beta production. J Exp Med. 1991, 173: 209-219. 10.1084/jem.173.1.209.
Jallow M, Teo YY, Small KS, Rockett KA, Deloukas P, Clark TG, Kivinen K, Bojang KA, Conway DJ, Pinder M, Sirugo G, Sisay-Joof F, Usen S, Auburn S, Bumpstead SJ, Campino S, Coffey A, Dunham A, Fry AE, Green A, Gwilliam R, Hunt SE, Inouye M, Jeffreys AE, Mendy A, Palotie A, Potter S, Ragoussis J, Rogers J, Rowlands K, Somaskantharajah E, Whittaker P, Widden C, Donnelly P, Howie B, Marchini J, Morris A, Sanjoaquin M, Achidi EA, Agbenyega T, Allen A, Amodu O, Corran P, Djimde A, Dolo A, Doumbo OK, Drakeley C, Dunstan S, Evans J, Farrar J, Fernando D, Hien TT, Horstmann RD, Ibrahim M, Karunaweera N, Kokwaro G, Koram KA, Lemnge M, Makani J, Marsh K, Michon P, Modiano D, Molyneux ME, Mueller I, Parker M, Peshu N, Plowe CV, Puijalon O, Reeder J, Reyburn H, Riley EM, Sakuntabhai A, Singhasivanon P, Sirima S, Tall A, Taylor TE, Thera M, Troye-Blomberg M, Williams TN, Wilson M, Kwiatkowski DP: Genome-wide and fine-resolution association analysis of malaria in West Africa. Nat Genet. 2009
Ozaki K, Inoue K, Sato H, Iida A, Ohnishi Y, Sekine A, Sato H, Odashiro K, Nobuyoshi M, Hori M, Nakamura Y, Tanaka T: Functional variation in LGALS2 confers risk of myocardial infarction and regulates lymphotoxin-alpha secretion in vitro. Nature. 2004, 429: 72-75. 10.1038/nature02502.
Acknowledgements
We thank Michael Good for his critical review of the manuscript and helpful comments. We thank Ferryanto Chalfein, Prayoga, Daud Rumere, Roesmini, Yoshi Elvi for technical and logistical assistance; Mitra Masyarakat Hospital staff for clinical support; Mauritz Okeseray, Jeanne Rini, Paulus Sugiarto and Lembaga Pengembangan Masyarakat Amungme Kamoro for support in Timika; and Mushtaq Hassanali, Dennis Manyenga (deceased), Gustav Moyo, Stella Stanslaus, Bernard John, Sofia Mbangukura and Ann Shumbusho for clinical, technical and logistical assistance in Dar es Salaam. We also thank Leanne Morrison and Dennis Moss for provision of EBV-transformed cell lines.
This work was supported by grants from the National Institutes of Health (AI55982 and AI041764), Australian NHMRC Programme Grants (290208 and 496600), the US V. A. Research Service, Australian Postgraduate Award to LMR, Australian NHMRC Fellowships to CRE, NMA and GM, Wellcome Trust Career Development Award to RNP and the Tudor Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
LMR performed experiments, analysed data and drafted the manuscript, EK, DAL, ET, EDM, TH, KP, ZZZ, FLR, YZ, KMS, LL, FHA, AH, ACS, TW and ES collected and processed samples, and performed experiments, DLG, MRH, RNP, JBW, GWM, NMA and CRE designed experiments analysed data and wrote the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Randall, L.M., Kenangalem, E., Lampah, D.A. et al. A study of the TNF/LTA/LTB locus and susceptibility to severe malaria in highland papuan children and adults. Malar J 9, 302 (2010). https://doi.org/10.1186/1475-2875-9-302
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2875-9-302