Abstract
Background
The antimalarial drug atovaquone specifically targets Plasmodium falciparum cytochrome b (Pfcytb), a mitochondrial gene with uniparental inheritance. Cases of resistance to atovaquone associated with mutant Pfcytb have been reported, justifying efforts to better document the natural polymorphism of this gene. To this end, a large molecular survey was conducted in several malaria endemic areas where atovaquone was not yet in regular use.
Methods
The polymorphism of the Pfcytb was analysed by direct sequencing of PCR products corresponding to the full length coding region. Sequence was generated for 671 isolates originating from three continents: Africa (Senegal, Ivory Coast, Central African Republic and Madagascar), Asia (Cambodia) and South America (French Guiana).
Results
Overall, 11 polymorphic sites were observed, of which eight were novel mutations. There was a large disparity in the geographic distribution of the mutants. All isolates from Senegal, Central African Republic and Madagascar displayed a Camp/3D7 wild type Pfcytb sequence, as did most samples originating from Cambodia and Ivory Coast. One synonymous (t759a at codon V253V) and two non-synonymous (t553g and a581g at codons F185V and H194R, respectively) singletons were detected in Ivory Coast. Likewise, two synonymous (a126t and c793t at codons -T42T and L265L, respectively) singletons were observed in Cambodia. In contrast, seven mutated sites, affecting seven codons and defining four mutant haplotypes were observed in French Guiana. The wild type allele was observed in only 14% of the French Guiana isolates. The synonymous c688t mutation at position L230L was highly prevalent; the most frequent allele was the c688t single mutant, observed in 84% of the isolates. The other alleles were singletons (a126t/a165c, a4g/a20t/a1024c and a20t/t341c/c688t corresponding to T42T/S55S, N2D/N71I/I342L, N71I/L114S/L230L, respectively" please replace with ' corresponding to T42T/S55S, N2D/N71I/I342L and N71I/L114S/L230L, respectively). The codon 268 polymorphisms associated with atovaquone resistance were not observed in the panel the isolates studied. Overall, the wild type PfCYTb protein isoform was highly predominant in all study areas, including French Guiana, suggesting stringent functional constraints.
Conclusion
These data along with previously identified Pfcytb field polymorphisms indicate a clustering of molecular signatures, suggesting different ancestral types in South America and other continents. The absence of mutations associated with most atovaquone-proguanil clinical failures indicates that the atovaquone-proguanil association is an interesting treatment option in the study areas.
Similar content being viewed by others
Background
A high rate of treatment failure for commonly anti-malarial drugs used in Plasmodium falciparum infections has been reported in numerous endemic areas. The recommended treatment policy is now to use drug combinations [1]. The atovaquone-proguanil (AP) drug combination, distributed under the trade name of Malarone®, is one of the treatment and prophylaxis options. AP has a high cure rate, limited mild side effects [2] and proved efficacious against multi-drug resistant P. falciparum malaria [3, 4]. Atovaquone (a hydroxy-naphthoquinone) is a potent inhibitor of the cytochrome bc1 (cytbc1) complex [5–8], a key respiratory enzyme from the mitochondrial membrane, while proguanil (an isopropylbiguanide) inhibits the plasmodial dihydrofolate reductase [4, 9, 10]. The association synergizes to collapse the mitochondrial membrane [8, 11].
Plasmodium falciparum in vitro resistance to atovaquone has been associated with specific point mutations in the cytochrome b gene (Pfcytb) in the region spanning codons 271–284 [7, 12]. A high frequency of recrudescence was observed in patients receiving atovaquone as a single drug therapy against P. falciparum [13, 14]. A Y268S point mutation in the Pfcytb gene, distinct from the mutations observed in the lines selected in vitro for atovaquone resistance, was detected in the recrudescing parasites [7]. Codon 268 polymorphism was used as marker for a molecular surveillance of atovaquone-proguanil resistance [15–19]. AP treatment failures were increasingly reported a few years after its introduction, with recrudescing parasites presenting a markedly increased IC50 for atovaquone [15, 20, 21]. In most cases, recrudescence was associated with a mutant 268 codon, either a Y268S [3, 12, 20, 22–26], a Y268N [15] or a Y268C mutation [21]. However, the presence of a mutant 268 codon was not observed in all cases of AP failure [21, 27].
Along with the key issue of emergence and spreading of polymorphisms conferring atovaquone resistance, analysis of Pfcytb field diversity presents an interest in population genetics [28–31]. Indeed, the cytb gene is encoded by the mitochondrial DNA and as a consequence, is of uniparental inheritance and under quite different evolution constraints compared to nuclear genes [32, 33]. In particular, interallelic recombination is not possible and polymorphisms such as base substitutions or insertions may accumulate over time. In mammals, the cytb locus displays an approximately 10-fold higher mutation rate than nuclear genes [34]. A rapid mutation rate (one mutation in 105 parasites) was reported for Pfcytb [35], but 100–1,000 lower rates were described subsequently [7].
Sequence polymorphism of the near to full gene sequence has been explored in laboratory [7, 36] and in vitro resistant isolates [7, 12]. It has also been looked for in cases of treatment failures from several countries, mostly African countries [7, 12, 15, 21, 22, 24–26]. Systematic analysis of field polymorphism in African settings [37, 38] and in isolates from the Thai Myanmar border [39] essentially focused on the region coding for the atovaquone-binding site. Full gene sequence analysis of field samples has been restricted to isolates from few patients in India [36] and from patients returning to France from West Africa, Central Africa or the Indian Ocean [21, 25, 40]. This provides an interesting picture of the overall polymorphism of the gene, but little clues on possible population signatures related to this gene. Sequence analysis of mitochondrial DNA from 100 independent isolates collected worlwide provided evidence for geographical clustering [29, 30].
To further document Pfcytb population polymorphism, 671 isolates from six different areas: Africa (Senegal, Ivory Coast, Central African Republic, and Madagascar), Asia (Cambodia) and South America (French Guiana) were AP had not been in regular use were analysed. This identified numerous novel, low frequency polymorphisms in Africa and Cambodia, most of which were country-specific, together with a high frequency signature that was specific for the South American isolates. None of the polymorphisms previously associated with atovaquone resistance in vitro or AP treatment failure was observed in this panel of isolates.
Methods
Study-sites and sample collection
Isolates were collected during the drug susceptibility surveillance programme conducted by the reference laboratory based in French Guiana, as such they were exempt from consent. Additional isolates originating from Senegal [41], Madagascar, Cambodia [42], Ivory Coast [43] and Central African Republic [44] were collected from patients recruited at home or at health centres during regular control surveys. Informed consent was obtained for these studies.
Blood samples were collected in each country from patients with mild malaria in the years 2000–2003, except in Central African Republic where blood was collected in the year 2004. After blood smear analysis, patients with mixed species infections were excluded. Only patients with positive slides for P. falciparum were included (Senegal : Sn, N = 45 ; Madagascar : Mg, N = 192 ; Ivory Coast : IC, N = 44 ; Cambodia : Kh, N = 179 ; French Guiana : FG, N = 160 ; Central African Republic : RCA, N = 51). Blood was stored frozen before being processed for genomic DNA isolation and amplification.
DNA extraction and Polymerase Chain Reaction (PCR) amplification
Parasite DNA was extracted from frozen blood aliquots using the phenol/chloroform method [45]. Amplifications were performed in 50 μL final reaction volume containing DNA template, 1 μM each primer, 200 μM each dNTP, 1.75 mM MgCl2 and 2.5 U Taq polymerase (Promega) using a Mastercycler Gradient 5331, Eppendorf. The primers were designed to amplify the full length Pfcytb gene, using as reference sequence the atovaquone-sensitive P. falciparum 3D7 clone (accession No AY282930), and carried an extension sequence (bold) that was used as sequencing primer as well: cytb1-sense (ctcgaggaattcggatcc tatgaacttttactctattaatt) and cytb2-antisense (tctagaaagcttggatcc tatatgtttgcttgggagct). The PCR amplification conditions were: 1 cycle denaturation at 94°C for 3 min, followed by 5 cycles [94°C for 30 sec, 56°C for 90 sec, 65°C for 150 sec] and 35 cycles [94°C for 10 sec, 53°C for 90 sec, 65°C for 150 sec]. A final extension was done at 65°C during 15 min. The isolates from Ivory Coast were amplified using the primers CYTb1 and CYTb2 [12].
Direct sequencing of PCR products
The PCR products were purified using a P-100 Gel Fine solution (Bio-Rad) and Multiscreen MAVN45 kit system (Millipore). The amount of PCR product (1131 bp) was estimated on a 1.2% agarose gel. Sequencing reactions were performed on both strands using the extension primers (sequence in bold indicated above) and internal primers using ABI Prism BigDye Terminator chemistry. Sequencing reactions conditions were as follows: 1 cycle at 96°C for 60 sec, followed by 25 cycles [96°C for 10 sec, 50°C for 5 sec, 60°C for 4 min]. The product was ethanol precipitated and washed with 70% Ethanol. The pellets were resuspended in 10 μL 0.3 mM EDTA and sequenced using an ABI PRISM 3100 Genetic analyzer (Applied Biosystems).
The isolates from Ivory Coast were sequenced on both strands with the internal primers PfCYTB33 (5'atttatgatatttattgtaactgc) and PfCYTB4R (5'agttggttaaacttctttgttctgc), covering codons 122 to 294 (i.e. encompassing the binding site of atovaquone).
Data analysis
Sequence analysis was done using Phred Phrap consed package [46]. Sequences with segments ≥ 1000 bp called with a quality over 20 per base were retained. Only unambiguous single nucleotide polymorphisms (SNPs) were considered. Sequences of insufficient quality were either resequenced or rejected. The sequence assembly was done with the Seqscape software v.2.0. (Applied Biosystems).
Results
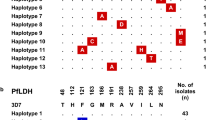
Among the 671 isolates, a full length Pfcytb sequence was successfully established for 576 isolates and partial sequence was obtained for 95 isolates (Figure 1). Overall, 11 polymorphic sites were observed (Figure 1), of which eight were novel mutations compared with published data (Figure 2).
Individual Pfcytb polymorphisms and allelic types observed in the study areas. WT : wild type ; * non synonymous mutation; ° synonymous mutation. The polymorphic sites observed in this study are shown by codon (numbers refer to positions in the protein sequence). Amino acids are indicated with single letter code and in capital, the nucleotide changes are indicated below (bold). Area-specific mutations are coloured in brown, blue and orange for Ivory Coast, Cambodia and French Guiana, respectively. In grey, T42T, the only mutation found common between two settings.
Compilation of the polymorphic sites of the Pfcytb gene in field isolates (in blue) and in lines selected for atovaquone-resistance in vitro (in yellow). The figure does not include silent mutations, which were not described [36] or whose nucleotide numbering does not match with the reported reference sequences [37, 40]. Numbering indicated refers to data reported from 1-Korzinczky et al [7]; 2-Schwöbel et al. [12]; 3-Sharma et al. [36]; 4-Musset et al. [25]; 5-Fivelman et al. [15]; 6-Berry et al. [40]; 7-Happi et al. [38]; 8-Kuhn et al. [24]; 9-Färnert et al. [23]; 10-David et al. [22]; 11-Schwartz et al. [26]; 12-Musset et al. [21]; 13-Joy et al. [30]; 14-Conway et al. [29]; 15-Legrand et al. [20]. # Single Nucleotide Polymorphism found in this study; * Musset personal communication; ** codon 282 is GTA : nucleotide 846 is an A and not a C as quoted by Berry et al [40], we assume that the T to C mutation affects nucleotide 845
All isolates from Senegal, Central African Republic and Madagascar had the same sequence, which was identical to the Camp/3D7 reference sequence. One synonymous point mutation and two coding mutations were observed in the set of isolates from Ivory Coast (Figure 1). None of these had been published previously and each was observed in a single isolate.
In Cambodia, two synonymous mutations (T42T and L265L) were observed (Figure 1). Each was detected in one isolate, the remaining 177 isolates harboured the Camp/3D7 reference Pfcytb gene sequence.
The French Guiana samples presented the largest polymorphism, with seven mutated sites affecting seven codons. Five alleles were observed (Figure 1). Unlike the other settings where alleles had a single mutated position, three out of the five alleles from French Guiana were multiple mutants with two (allele FG2) or three (alleles FG3 and FG4) mutated codons. The Camp/3D7 reference allele was present in 22 out of 160 isolates (14%). Allele FG1, which carried a silent L230L mutation, was highly predominant, accounting for 135 of 160 (84%) isolates (Figures 1 and 3). The same mutation was associated with additional SNPs in allele FG4, which therefore probably derive from FG1. Allele FG4 which carried additional mutations N7I and L114S, was observed only once (frequency 0.6%). The N7I polymorphism was detected in allele FG3 as well, but in this case it was associated with N2D and I342L (Figure 1).
Geographic distribution of the Pfcytb gene polymorphism studied here. Country's code: FG: French Guiana, Sn: Senegal, IC: Ivory Coast, RCA: Central Africa, Mg: Madagascar, Kh: Cambodia. Allelic frequency in each site is depicted in colour coded pie charts: brown, blue and orange referring to Ivory Coast, Cambodia and French Guiana, respectively. The wild type allele (WT) is indicated in white.
Discussion
In the panel of isolates studied here, 11 polymorphic sites were observed, resulting in 11 codons displaying a single point mutation. Ten distinct alleles could be identified, as shown in Figure 1. Apart from the a126t and c688t silent mutations at codon T42T and L230L, respectively, and a581g non synonymous mutation at codon H194R, all mutations observed here were novel (Figures 1 and 2). Five synonymous and six non-synonymous mutations were detected. The observed ratio of synonymous to non synonymous mutations is in line with four synonymous and five non synonymous mutations detected in 270 full gene sequences of isolates from West Africa and the Indian Ocean [25] and four synonymous and six non synonymous mutations in 14 isolates from India [36], but lower than the three to one ratio observed in Gabon [37] and the nine to three ratio reported for 135 isolates from West and Central Africa [40]. The compilation of all Pfcytb field polymorphisms identified so far (Figure 2) highlights 50 polymorphic sites, affecting 45 codons, with 30 mutant amino acid residues.
There was a clear geographical heterogeneity in the level and type of polymorphism (Figure 3). The Camp/3D7 type was the single allelic form detected in three African settings studied here (Madagascar, Senegal and Central African Republic). Similar observations were made in Ethiopia [37]. In Ivory Coast, the same allele was observed in 93% of the patients, a frequency similar to the 90% prevalence observed in Gabon [37], or in travellers returning to France from West and Central Africa [40]. As observed in other African endemic settings [37], the mutant alleles were single mutants and each mutant had a low frequency, being observed in one or two isolates. A similar low frequency of single mutant alleles has been observed in the panel of isolates from patients returning from travel to West and Central Africa and the Indian Ocean [25, 40].
In Cambodia, the Camp/3D7 wild type allele was also highly predominant, consistent with recent sequence data of the atovaquone binding site in patients from the Thai/Myanmar border [39]. The two mutations observed were both synonymous and detected at low frequency.
French Guiana showed a quite different profile, and was the most polymorphic of the six countries explored. Seven of 11 mutant sites were observed in the set of isolates from French Guiana. Furthermore, this area was the only one where multiple mutant alleles were detected. Importantly, the Camp/3D7 wild type allele, which was dominant in the other settings was observed with a 14% frequency only, while the dominant allele (84% frequency) was a single, silent L230L mutant. This was in accordance with data from other localities from South America [29, 30]. Triple mutants, most probably originating from the L230L parent for FG4 isolate, were observed along with a double mutant possibly derived from the Camp/3D7 type. Thus, the French Guiana parasite population had a Pfcytb pattern dissimilar from the African and Cambodian settings. This was consistent with data from others [29, 30]. A specific, high frequency geographical signature seems to prevail in India as well, where 13 or 14 isolates carried a N2N F3Y Y4Y haplotype [36]. However, polymorphism in French Guiana was larger than in India, where three allelic forms have been reported. Thus, of all geographical areas studied so far, French Guiana presents the largest Pfcytb gene polymorphism. This is interpreted as a consequence of its population structure, with hypoendemic, isolated foci that are propitious to genetic drift [47].
Pfcytb nucleotide polymorphism was larger than the deduced protein sequence. All three alleles from Cambodia coded for the same, wild type protein sequence. In French Guiana the wild type deduced protein sequence accounted for 158 of 160 alleles. Thus in all geographic regions, the wild type protein sequence was the highly dominant if not the only predicted isoform. Altogether, these data point to an elevated mutation rate of the locus, with albeit a large dominance of the wild type protein sequence, probably indicating its optimal fitness. At the nucleotide level, interesting geographic clustering of molecular signatures were observed (Figure 4), suggesting different ancestral types in French Guiana (and in South America) as well as in India [36]. Specificities of the parasite population in India is also suggested by polymorphism of the Pfcrt locus [48]. The data reported here further support the evidence that the present parasite population from South America differs from the parasites from Africa, regarding surface antigens [49], loci such as Pfcg2 [31], Pfcrt [50–52], Pfdhfr and Pfdhps [51] and numerous genome-wide scattered SNPs [53]. The large Pfcytb polymorphism together with the observation that the var gene repertoire in Brazilian isolates is small and highly redundant unlike in Africa and Southeast Asia [54], raises the question of the structuring of the South American P. falciparum population.
Phylogenetic relationship among 20 Pfcytb alleles from various geographical origin. The tree was estimated using the Phylip DNAdist program and the neighbour-joining method (100 replications), and drawn using Drawtree software. Among the various Pfcytb haplotypes previously published, only those corresponding to a natural polymorphism of the gene (i.e. not selected under atovaquone pressure) were considered for the phylogenetic analysis. The 3D7 sequence >gi|31789515|gb|AY282930 was used as reference. The haplotypes were as follows: Nga1: G757A [40]; Nga2: T802A, Nga3: C796A [38]; Sen1: G160C, Sen2: G209A, Mali1: T918A, Mali2: T1086A, Ben1: A126T, Com: C234T, Burk1: C1059T, RCA1: C90T [21]; Kh1: A126T, Kh2: C793T, FG1: C688T, FG2:A126T/A165C, FG3:A4G/A20T/A1024C; FG4:A20T/T341C/C688T (this paper); Ind1: T273C/A377T/C494G/A861T/T862A/T929A, Ind2: C6T/T8A/C12T/A275G, Ind3: C6T/T8A/C12T/A275G/T977C [36]. Codes used for countries: Senegal: Sen, Comoro islands: Com, India: Ind, Central Africa: RCA, Mali, Nigeria: Nga, French Guiana: FG, Benin: Ben, Burkina Faso: Burk, Kh: Cambodia.
None of the field polymorphism observed here or in other settings concerned the residues that are mutated in lines selected in vitro for atovaquone resistance [5, 7, 12]. Furthermore, no mutant Y268S, Y268C and Y268N Pfcytb, selected under atovaquone or AP pressure [3, 7, 12, 15, 21, 22, 24–26] was detected in any of the settings explored here. The 268N mutant, which has been observed with a 4.5% frequency in Nigeria in the absence of AP pressure [38] was not detected. This mutation has been reported so far essentially in Nigeria [15, 38] and, as discussed by Happi et al [38] may have arisen under pressure by related drugs. The Y268S polymorphism was not detected in the isolates from French Guiana studied here, which were collected before implementation of AP as prophylaxis and as second line treatment in 2002. It was however observed one year after implementation in a second line AP-treatment failure [20], further substantiating the interpretation of its selection during AP treatment.
Conclusion
Overall, the available data indicate an elevated mutation frequency of the Pfcytb locus, with multiple polymorphic sites, and in some cases more than one polymorphism per site, consistent with the elevated mutation frequency of mitochondrial genes [34, 35]. The geographical clustering observed here adds further support to the evidence that the parasite population from South America differs from the parasites from Africa. Importantly, the absence of a mutated codon 268 in all settings investigated here indicates that AP remains an interesting treatment option for these areas. In view of the high mutation rate and of the rapid selection of mutants under AP pressure, careful surveillance of emerging Pfcytb mutants and resistance to atovaquone used as prophylaxis in travellers or as treatment is warranted.
References
WHO Guidelines for the treatment of malaria. 2006, [http://www.who.int/malaria/diagnosisandtreatment.html]
Looareesuwan S, Wilairatana P, Chalermarut K, Rattanapong Y, Canfield CJ, Hutchinson DB: Efficacy and safety of atovaquone/proguanil compared with mefloquine for treatment of acute Plasmodium falciparum malaria in Thailand. Am J Trop Med Hyg. 1999, 60: 526-532.
Srivastava IK, Morrisey JM, Darrouzet E, Daldal F, Vaidya AB: Resistance mutations reveal the atovaquone-binding domain of cytochrome b in malaria parasites. Mol Microbiol. 1999, 33: 704-711. 10.1046/j.1365-2958.1999.01515.x.
Vaidya AB, Mather MW: Atovaquone resistance in malaria parasites. Drug Resist Updat. 2000, 3: 283-287. 10.1054/drup.2000.0157.
Kessl JJ, Ha KH, Merritt AK, Lange BB, Hill P, Meunier B, Meshnick SR, Trumpower BL: Cytochrome b mutations that modify the ubiquinol-binding pocket of the cytochrome bc1 complex and confer anti-malarial drug resistance in Saccharomyces cerevisiae. J Biol Chem. 2005, 280: 17142-17148. 10.1074/jbc.M500388200.
Kessl JJ, Lange BB, Merbitz-Zahradnik T, Zwicker K, Hill P, Meunier B, Palsdottir H, Hunte C, Meshnick S, Trumpower BL: Molecular basis for atovaquone binding to the cytochrome bc1 complex. J Biol Chem. 2003, 278: 31312-31318. 10.1074/jbc.M304042200.
Korsinczky M, Chen N, Kotecka B, Saul A, Rieckmann K, Cheng Q: Mutations in Plasmodium falciparum cytochrome b that are associated with atovaquone resistance are located at a putative drug-binding site. Antimicrob Agents Chemother. 2000, 44: 2100-2108. 10.1128/AAC.44.8.2100-2108.2000.
Painter HJ, Morrisey JM, Mather MW, Vaidya AB: Specific role of mitochondrial electron transport in blood-stage Plasmodium falciparum. Nature. 2007, 446: 88-91. 10.1038/nature05572.
Srivastava IK, Rottenberg H, Vaidya AB: Atovaquone, a broad spectrum antiparasitic drug, collapses mitochondrial membrane potential in a malarial parasite. J Biol Chem. 1997, 272: 3961-3966. 10.1074/jbc.272.52.33360.
Vaidya AB, Lashgari MS, Pologe LG, Morrisey J: Structural features of Plasmodium cytochrome b that may underlie susceptibility to 8-aminoquinolines and hydroxynaphthoquinones. Mol Biochem Parasitol. 1993, 58: 33-42. 10.1016/0166-6851(93)90088-F.
Srivastava IK, Vaidya AB: A mechanism for the synergistic antimalarial action of atovaquone and proguanil. Antimicrob Agents Chemother. 1999, 43: 1334-1339.
Schwobel B, Alifrangis M, Salanti A, Jelinek T: Different mutation patterns of atovaquone resistance to Plasmodium falciparum in vitro and in vivo: rapid detection of codon 268 polymorphisms in the cytochrome b as potential in vivo resistance marker. Malar J. 2003, 2: 5-10.1186/1475-2875-2-5.
Chiodini PL, Conlon CP, Hutchinson DB, Farquhar JA, Hall AP, Peto TE, Birley H, Warrell DA: Evaluation of atovaquone in the treatment of patients with uncomplicated Plasmodium falciparum malaria. J Antimicrob Chemother. 1995, 36: 1073-1078. 10.1093/jac/36.6.1073.
Looareesuwan S, Viravan C, Webster HK, Kyle DE, Hutchinson DB, Canfield CJ: Clinical studies of atovaquone, alone or in combination with other antimalarial drugs, for treatment of acute uncomplicated malaria in Thailand. Am J Trop Med Hyg. 1996, 54: 62-66.
Fivelman QL, Butcher GA, Adagu IS, Warhurst DC, Pasvol G: Malarone treatment failure and in vitro confirmation of resistance of Plasmodium falciparum isolate from Lagos, Nigeria. Malar J. 2002, 1: 1-10.1186/1475-2875-1-1.
Gil JP, Nogueira F, Stromberg-Norklit J, Lindberg J, Carrolo M, Casimiro C, Lopes D, Arez AP, Cravo PV, Rosario VE: Detection of atovaquone and Malarone resistance conferring mutations in Plasmodium falciparum cytochrome b gene (cytb). Mol Cell Probes. 2003, 17: 85-89. 10.1016/S0890-8508(03)00006-9.
Muehlen M, Schreiber J, Ehrhardt S, Otchwemah R, Jelinek T, Bienzle U, Mockenhaupt FP: Short communication: Prevalence of mutations associated with resistance to atovaquone and to the antifolate effect of proguanil in Plasmodium falciparum isolates from northern Ghana. Trop Med Int Health. 2004, 9: 361-363. 10.1111/j.1365-3156.2004.01201.x.
Pimentel S, Nogueira F, Benchimol C, Quinhentos V, Bom J, Varandas L, do Rosario V, Bernardino L: Detection of atovaquone-proguanil resistance conferring mutations in Plasmodium falciparum cytochrome b gene in Luanda, Angola. Malar J. 2006, 5: 30-10.1186/1475-2875-5-30.
Wichmann O, Muehlberger N, Jelinek T, Alifrangis M, Peyerl-Hoffmann G, Muhlen M, Grobusch MP, Gascon J, Matteelli A, Laferl H, Bisoffi Z, Ehrhardt S, Cuadros J, Hatz C, Gjorup I, McWhinney P, Beran J, da Cunha S, Schulze M, Kollaritsch H, Kern P, Fry G, Richter J: Screening for mutations related to atovaquone/proguanil resistance in treatment failures and other imported isolates of Plasmodium falciparum in Europe. J Infect Dis. 2004, 190: 1541-1546. 10.1086/424469.
Legrand E, Demar M, Volney B, Ekala MT, Quinternet M, Bouchier C, Fandeur T, Rogier C, Carme B, Puijalon O, Esterre P: First case of emergence of atovaquone resistance in Plasmodium falciparum during second-line atovaquone-proguanil treatment in South America. Antimicrob Agents Chemother. 2007, 51: 2280-2281. 10.1128/AAC.01532-06.
Musset L, Bouchaud O, Matheron S, Massias L, Le Bras J: Clinical atovaquone-proguanil resistance of Plasmodium falciparum associated with cytochrome b codon 268 mutations. Microbes Infect. 2006, 8: 2599-2604. 10.1016/j.micinf.2006.07.011.
David KP, Alifrangis M, Salanti A, Vestergaard LS, Ronn A, Bygbjerg IB: Atovaquone/proguanil resistance in Africa: a case report. Scand J Infect Dis. 2003, 35: 897-898.
Farnert A, Lindberg J, Gil P, Swedberg G, Berqvist Y, Thapar MM, Lindegardh N, Berezcky S, Bjorkman A: Evidence of Plasmodium falciparum malaria resistant to atovaquone and proguanil hydrochloride: case reports. Bmj. 2003, 326: 628-629. 10.1136/bmj.326.7390.628.
Kuhn S, Gill MJ, Kain KC: Emergence of atovaquone-proguanil resistance during treatment of Plasmodium falciparum malaria acquired by a non-immune north American traveller to west Africa. Am J Trop Med Hyg. 2005, 72: 407-409.
Musset L, Pradines B, Parzy D, Durand R, Bigot P, Le Bras J: Apparent absence of atovaquone/proguanil resistance in 477 Plasmodium falciparum isolates from untreated French travellers. J Antimicrob Chemother. 2006, 57: 110-115. 10.1093/jac/dki420.
Schwartz E, Bujanover S, Kain KC: Genetic confirmation of atovaquone-proguanil-resistant Plasmodium falciparum malaria acquired by a nonimmune traveler to East Africa. Clin Infect Dis. 2003, 37: 450-451. 10.1086/375599.
Wichmann O, Muehlen M, Gruss H, Mockenhaupt FP, Suttorp N, Jelinek T: Malarone treatment failure not associated with previously described mutations in the cytochrome b gene. Malar J. 2004, 3: 14-10.1186/1475-2875-3-14.
Conway DJ: Tracing the dawn of Plasmodium falciparum with mitochondrial genome sequences. Trends Genet. 2003, 19: 671-674. 10.1016/j.tig.2003.10.007.
Conway DJ, Fanello C, Lloyd JM, Al-Joubori BM, Baloch AH, Somanath SD, Roper C, Oduola AM, Mulder B, Povoa MM, Singh B, Thomas AW: Origin of Plasmodium falciparum malaria is traced by mitochondrial DNA. Mol Biochem Parasitol. 2000, 111: 163-171. 10.1016/S0166-6851(00)00313-3.
Joy DA, Feng X, Mu J, Furuya T, Chotivanich K, Krettli AU, Ho M, Wang A, White NJ, Suh E, Beerli P, Su XZ: Early origin and recent expansion of Plasmodium falciparum. Science. 2003, 300: 318-321. 10.1126/science.1081449.
Su X, Kirkman LA, Fujioka H, Wellems TE: Complex polymorphisms in an approximately 330 kDa protein are linked to chloroquine-resistant P. falciparum in Southeast Asia and Africa. Cell. 1997, 91: 593-603. 10.1016/S0092-8674(00)80447-X.
Creasey A, Mendis K, Carlton J, Williamson D, Wilson I, Carter R: Maternal inheritance of extrachromosomal DNA in malaria parasites. Mol Biochem Parasitol. 1994, 65: 95-98. 10.1016/0166-6851(94)90118-X.
Creasey AM, Ranford-Cartwright LC, Moore DJ, Williamson DH, Wilson RJ, Walliker D, Carter R: Uniparental inheritance of the mitochondrial gene cytochrome b in Plasmodium falciparum. Curr Genet. 1993, 23: 360-364. 10.1007/BF00310900.
Contamine V, Picard M: Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast. Microbiol Mol Biol Rev. 2000, 64: 281-315. 10.1128/MMBR.64.2.281-315.2000.
Gassis S, Rathod PK: Frequency of drug resistance in Plasmodium falciparum: a nonsynergistic combination of 5-fluoroorotate and atovaquone suppresses in vitro resistance. Antimicrob Agents Chemother. 1996, 40: 914-919.
Sharma I, Rawat DS, Pasha ST, Biswas S, Sharma YD: Complete nucleotide sequence of the 6 kb element and conserved cytochrome b gene sequences among Indian isolates of Plasmodium falciparum. Int J Parasitol. 2001, 31: 1107-1113. 10.1016/S0020-7519(01)00218-1.
Gebru T, Hailu A, Kremsner PG, Kun JF, Grobusch MP: Molecular surveillance of mutations in the cytochrome b gene of Plasmodium falciparum in Gabon and Ethiopia. Malar J. 2006, 5: 112-10.1186/1475-2875-5-112.
Happi CT, Gbotosho GO, Folarin OA, Milner D, Sarr O, Sowunmi A, Kyle DE, Milhous WK, Wirth DF, Oduola AM: Confirmation of emergence of mutations associated with atovaquone-proguanil resistance in unexposed Plasmodium falciparum isolates from Africa. Malar J. 2006, 5: 82-10.1186/1475-2875-5-82.
Naoshima-Ishibashi Y, Iwagami M, Kawazu S, Looareesuwan S, Kano S: Analyses of cytochrome b mutations in Plasmodium falciparum isolates in Thai-Myanmar border. Travel Med Infect Dis. 2007, 5: 132-134. 10.1016/j.tmaid.2006.07.002.
Berry A, Senescau A, Lelievre J, Benoit-Vical F, Fabre R, Marchou B, Magnaval JF: Prevalence of Plasmodium falciparum cytochrome b gene mutations in isolates imported from Africa, and implications for atovaquone resistance. Trans R Soc Trop Med Hyg. 2006, 100: 986-988. 10.1016/j.trstmh.2006.02.004.
Jambou R, Legrand E, Niang M, Khim N, Lim P, Volney B, Ekala MT, Bouchier C, Esterre P, Fandeur T, Mercereau-Puijalon O: Resistance of Plasmodium falciparum field isolates to in-vitro artemether and point mutations of the SERCA-type PfATPase6. Lancet. 2005, 366: 1960-1963. 10.1016/S0140-6736(05)67787-2.
Khim N, Bouchier C, Ekala MT, Incardona S, Lim P, Legrand E, Jambou R, Doung S, Puijalon OM, Fandeur T: Countrywide survey shows very high prevalence of Plasmodium falciparum multilocus resistance genotypes in Cambodia. Antimicrob Agents Chemother. 2005, 49: 3147-3152. 10.1128/AAC.49.8.3147-3152.2005.
Bogreau H, Renaud F, Bouchiba H, Durand P, Assi SB, Henry MC, Garnotel E, Pradines B, Fusai T, Wade B, Adehossi E, Parola P, Kamil MA, Puijalon O, Rogier C: Genetic diversity and structure of African Plasmodium falciparum populations in urban and rural areas. Am J Trop Med Hyg. 2006, 74: 953-959.
Menard D, Djalle D, Manirakiza A, Yapou F, Siadoua V, Sana S, Matsika-Claquin MD, Nestor M, Talarmin A: Drug-resistant malaria in Bangui, Central African Republic: an in vitro assessment. Am J Trop Med Hyg. 2005, 73: 239-243.
Contamin H, Fandeur T, Bonnefoy S, Skouri F, Ntoumi F, Mercereau-Puijalon O: PCR typing of field isolates of Plasmodium falciparum. J Clin Microbiol. 1995, 33: 944-951.
Ewing B, Hillier LD, Wendl MC, Green P: Base-calling of automated sequencer traces using Phred I. Accuracy Assessment. Genome Res. 1998, 8: 175-85.
Ariey F, Duchemin JB, Robert V: Metapopulation concepts applied to falciparum malaria and their impacts on the emergence and spread of chloroquine resistance. Infect Genet Evol. 2003, 2: 185-192. 10.1016/S1567-1348(02)00099-0.
Das A, Dash AP: Evolutionary paradigm of chloroquine-resistant malaria in India. Trends Parasitol. 2007, 23: 132-135. 10.1016/j.pt.2007.01.012.
Creasey A, Fenton B, Walker A, Thaithong S, Oliveira S, Mutambu S, Walliker D: Genetic diversity of Plasmodium falciparum shows geographical variation. Am J Trop Med Hyg. 1990, 42: 403-413.
Ariey F, Fandeur T, Durand R, Randrianarivelojosia M, Jambou R, Legrand E, Ekala MT, Bouchier C, Cojean S, Duchemin JB, Robert V, Le Bras J, Mercereau-Puijalon O: Invasion of Africa by a single pfcrt allele of South East Asian type. Malar J. 2006, 5: 34-10.1186/1475-2875-5-34.
Cortese JF, Caraballo A, Contreras CE, Plowe CV: Origin and dissemination of Plasmodium falciparum drug-resistance mutations in South America. J Infect Dis. 2002, 186: 999-1006. 10.1086/342946.
Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LM, Sidhu AB, Naude B, Deitsch KW, Su XZ, Wootton JC, Roepe PD, Wellems TE: Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000, 6: 861-871. 10.1016/S1097-2765(05)00077-8.
Mu J, Awadalla P, Duan J, McGee KM, Joy DA, McVean GA, Su XZ: Recombination hotspots and population structure in Plasmodium falciparum. PLoS Biol. 2005, 3: e335-10.1371/journal.pbio.0030335.
Albrecht L, Merino EF, Hoffmann EH, Ferreira MU, de Mattos Ferreira RG, Osakabe AL, Dalla Martha RC, Ramharter M, Durham AM, Ferreira JE, del Portillo HA, Wunderlich G: Extense variant gene family repertoire overlap in Western Amazon Plasmodium falciparum isolates. Mol Biochem Parasitol. 2006, 150: 157-165. 10.1016/j.molbiopara.2006.07.007.
Acknowledgements
We are indebted to the patients for their invaluable contribution in the study. We are grateful to the field teams involved in patient care and acknowledge field-based laboratory staff for technical assistance. We acknowledge for their technical support the whole team of PT1-Genopole, in particular Laurence Ma, Nora Zidane and Magali Tichit. Helpful comments on the manuscript were given by Dr Frederic Ariey. Financial supports were provided by Académie des Sciences (prix Louis D.), E.U. grant Resmalchip contract QLK2-CT20021-1503, Génopole (Pasteur Institute of Paris, France), FSP-RAI composante paludisme du Ministère des affaires étrangères.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
MTE did the sequencing of isolates except for Ivory Coast, was responsible for data collection, entry in the database and drafted the manuscript.
RJ (Senegal), MR (Madagascar), EL (French Guiana), DM (Republic Central Africa), SBA, MCH, CR (Ivory Coast) were responsible for sample collection and coordination of laboratory in the field.
NK, EL, MR, RJ, DM and CR performed all PCR amplifications and actively participated in sample collection.
TF established suitable protocols for Pfcytb amplification, conducted the phyologenetic study, participated in drafting the manuscript and was responsible for coordination of laboratory work in Cambodia.
CB was responsible for the sequencing procedure.
OMP conceived the study, helped for sequence analysis, drafted and revised the manuscript.
All authors read and gave the final approval of the version to be published.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Ekala, MT., Khim, N., Legrand, E. et al. Sequence analysis of Plasmodium falciparum cytochrome b in multiple geographic sites. Malar J 6, 164 (2007). https://doi.org/10.1186/1475-2875-6-164
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2875-6-164