Abstract
Background
Norvaline is an unusual non-proteinogenic branched-chain amino acid which has been of interest especially during the early enzymological studies on regulatory mutants of the branched-chain amino acid pathway in Serratia marcescens. Only recently norvaline and other modified amino acids of the branched-chain amino acid synthesis pathway got attention again when they were found to be incorporated in minor amounts in heterologous proteins with a high leucine or methionine content. Earlier experiments have convincingly shown that norvaline and norleucine are formed from pyruvate being an alternative substrate of α-isopropylmalate synthase, however so far norvaline accumulation was not shown to occur in non-recombinant strains of E. coli.
Results
Here we show that oxygen limitation causes norvaline accumulation in E. coli K-12 W3110 during grow in glucose-based mineral salt medium. Norvaline accumulates immediately after a shift to oxygen limitation at high glucose concentration. On the contrary free norvaline is not accumulated in E. coli W3110 in aerobic cultures. The analysis of medium components, supported by transcriptomic studies proposes a purely metabolic overflow mechanism from pyruvate into the branched chain amino acid synthesis pathway, which is further supported by the significant accumulation of pyruvate after the oxygen downshift. The results indicate overflow metabolism from pyruvate as necessary and sufficient, but deregulation of the branched chain amino acid pathway may be an additional modulating parameter.
Conclusion
Norvaline synthesis has been so far mainly related to an imbalance of the synthesis of the branched chain amino acids under conditions were pyruvate level is high. Here we show that simply a downshift of oxygen is sufficient to cause norvaline accumulation at a high glucose concentration as a consequence of the accumulation of pyruvate and its direct chain elongation over α-ketobutyrate and α-ketovalerate.
Although the flux to norvaline is low, millimolar concentrations are accumulated in the cultivation broth, which is far above the level which has been discussed for being relevant for misincorporation of norvaline into recombinant proteins. Therefore we believe that our finding is relevant for recombinant protein production but also may even have implications for the physiology of E. coli under oxygen limitation in general.
Similar content being viewed by others
Background
Norvaline belongs to the group of non-usual amino acid analogs that may be formed under certain circumstances as byproducts of the branched-chain amino acid biosynthetic pathway in E. coli and other Gram-negative microorganisms. These amino acids can accumulate and are secreted under certain cultivation conditions.
Historically, norvaline has been reported in one example to be a natural component of an antifungal peptide produced by Bacillus subtilis [1]. Later it was found as a side product in isoleucine overproducing regulatory mutants of Serratia marcescens which initiated a number of physiological studies by Chibata and colleagues [2–7]. Recently, the formation of modified branched chain amino acids has got increasing attention as these amino acids appeared to be incorporated in certain recombinant proteins produced in E. coli, e.g. β-methylnorleucine into a recombinant hirudin [8–10], norleucine into recombinant human brain derived neurotrophic factor [11], interleukin 2 [12–14], and bovine somatotropin [15], and norvaline into recombinant hemoglobin [16].
Incorporation of the modified amino acids into proteins occurs over mis-aminoacylation of tRNAs; namely of leucine-tRNA by norvaline, methionine-tRNA by norleucine [11–23] and to a low probability by norvaline, and ile-tRNA by β-methylnorleucine.
The incorporation of these unusual amino acids in recombinant proteins has been assigned to conditions which derepress the branched-chain amino acid pathway, which can occur due to strong expression of leucine-rich proteins [14]. Subsequently derepression of the pathway occurs by the low concentration of the proteinogenic amino acid leucine.
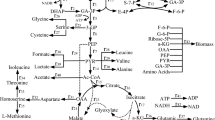
The synthesis of the modified amino acids of the branched-chain amino acid synthesis pathway derives from the isoleucine route which starts with α-ketobutyrate and their synthesis is caused by the low specificity of the enzymes of the branched-chain amino acid synthesis pathway (including the gene products of the ilv and leu operons) for α-keto acids. A probable scheme of the synthesis of the modified amino acids has been published by Apostol et al. [16] and is shown for norvaline in Figure 1.
Schematic view of the branched chain amino acid pathway with the proposed pathway for norvaline synthesis from pyruvate in relation with glycolysis and the tricarbonic acid cycle in E. coli . Overflow metabolites and anaerobic routes are included. For simplification some metabolic pathways with more steps are illustrated by an interrupted line. Numbers relate to the amount of enzymatic reactions. Enzymes are indicated by their gene names.
The branched-chain amino acid pathway for the synthesis of the three amino acids isoleucine, valine and leucine is closely interconnected to the glycolytic pathway. The major glycolysis-based metabolic intermediates involved are pyruvate and acetyl-CoA. α-ketobutyrate which is the substrate for the isoleucine pathway is synthesized from oxaloacetate over a number of intermediates, including L-aspartate, homoserine, and threonine. However, as a shorter and probable alternative for accumulation of α-ketobutyrate during conditions where the modified amino acids accumulate transiently, the direct extension of the carbon chain of pyruvate by the enzymes of the leu operon was proposed by Bogosian et al. [15] supported by kinetic data for α-isopropylmatate synthase from Salmonella typhimurium [24] and Serratia marcescens [2].
The specific feature of the branched amino acid pathway which has made this route interesting for a number of molecular-physiological and biochemistry studies is (i) the interactive metabolic feed-back regulation of synthetic enzymes by accumulation of one or more of the amino acids. In consequence amino acid starvation responses can be provoked by over-accumulation of one of the amino acids, which leads to suppression of the synthesis of the others. A well established example is the induction of the stringent response in E. coli K-12 strains by addition of valine which results in isoleucine starvation [25, 26]. (ii) The expression levels of the enzymes of the branched-chain amino acid pathway are regulated in dependence on the cellular concentration of each of the branched chain amino acids by the interactive repression of mRNA synthesis of the operons of the metabolic pathway by attenuation. (iii) Finally, a specific characteristic of the enzymes of this pathway is their low substrate specificity. Most of the enzymes can accept other than the main α-ketoacids as substrate. As a result, also other modified amino acids can be synthesized. Norvaline and norleucine directly diverge from α-ketobutyrate by chain elongation through the action of α-isopropylmalate synthase (α-IPMS), α-isoprolylmalate isomerase, β-isoprolylmalate dehydrogenase and following transamination. For this reaction accumulation of α-ketobutyrate is a prerequisite, because the affinity of α-IPMS for α-ketobutyrate is an order of magnitude higher compared to its natural substrate α-ketoisovalerate [2] as detected for S. marcescens. No enzymatic data are available to our knowledge for the corresponding E. coli enzymes.
This possibility of the enzymes encoded by the leuABCD operon to use other substrates suggests a direct possibility for chain elongation from pyruvate (3C) over α-ketobutyrate (4C) and α-ketovalerate (5C) to α-ketocaproate (6C). This hypothesis is further supported by experimental data from Apostol et al. [16]. In their study the first response to hemoglobin induction is a decrease of the leucine pool and the accumulation of pyruvate, which results in the immediate increase of norvaline, followed by a later accumulation of norleucine.
In the actual study we show by metabolic shift experiments that norvaline is not accumulated in wild type E. coli K-12 strains during aerobic cultivation on mineral salt medium, but only after a switch from aerobic to anaerobic conditions at a high glucose concentration. Our results support the model proposing that pyruvate is directly converted to α-ketobutyrate, which indicates that also conditions which lead to accumulation of pyruvate by natural responses may cause norvaline synthesis. The results also provide evidence that norvaline synthesis after an oxygen downshift is not related to induction or derepression of the enzymes of the branched chain amino acids, but rather to a metabolic overflow reaction.
Results
Earlier it has been suggested that norvaline synthesis may be triggered by conditions which lead to accumulation of pyruvate [16]. It is well known that pyruvate strongly increases in E. coli during unlimited growth on glucose if the environment is shifted from aerobic to anaerobic conditions. However, it has not been tested so far whether such a shift also stimulates norvaline synthesis.
As the norvaline level in aerobically grown cells is below the detection limit (Fig. 2A) and difficult to analyze at low cell densities (own unpublished results), we performed cultivations at high cell densities. Therefore E. coli W3110 was cultivated on mineral salt medium with an initial glucose concentration of 40 g L-1. The culture was grown up to an OD600 of about 40 (about 15 g L-1 cell dry weight) when oxygen limitation was initiated by a step-decrease of the agitation rate, resulting in a lower oxygen transfer rate and consequently the dissolved oxygen tension (DOT) decreased immediately to zero. The amount of glucose was kept non-limiting by constant feeding of a highly concentrated glucose solution in a fed-batch mode. As a control an aerobic cultivation was performed in the same way, however with a glucose-limited feeding to avoid oxygen limitation. As is seen from figure 3, the typical anaerobic metabolites of the mixed acid fermentation of E. coli accumulate after the downshift of the stirrer rate including acetate, formate, succinate, lactate, and ethanol indicating that the culture responds by anaerobic metabolism despite the continuous supply of air.
High cell density cultures of E. coli W3110 without (A, C) and with a stirrer downshift resulting in oxygen limited culture (B, D). The graphs show the data for cell dry weight (g L-1), dissolved oxygen tension (DOT), glucose concentration (A, B) and the anaerobic metabolites acetate, lactate, formate, ethanol, succinate (C, D, symbols shown in C). Anaerobic conditions in graphs (B, D) were caused by a downshift of the stirrer speed whereas the aeration rate was kept constant, as described in the Material and Methods section. The glucose feed was started at 0 hours. The stirrer downshift was performed at -10 min.
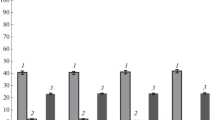
As expected, aside from a decrease of the cell growth, the anaerobic shift also caused accumulation of pyruvate. Pyruvate is a central metabolic compound for many pathways. Therefore one might hypothesisze that such a significant accumulation of pyruvate affects not only the flow into the typical anaerobic metabolic pathways but also into the pyruvate-connected routes for amino acid synthesis. To investigate this, we analyzed the concentrations of free amino acids in both cultivation types, with a downshift of the stirrer rate and without (Fig. 4). Interestingly, in agreement with our hypothesis we detected a significant increase in certain amino acids after the switch to anaerobic growth conditions especially in pathways which are closely connected to pyruvate, such as alanine, valine, and somewhat less significantly for leucine. Also glutamine and aspartate showed a significant increase, and asparagine increased with a similar kinetics as aspartate, but the concentration remained three orders of magnitude lower (μM range). In contrast isoleucine did not increase (see Fig. 4), and similarly most of the other amino acids, which are not directly related to pyruvate, did not increase in their concentrations (not shown).
Concentrations of free amino acids and pyruvate during fed-batch cultivations of E. coli W3110 without (as control, open squares) and with a stirrer downshift resulting in an oxygen limited culture (closed triangles). The analysis was performed from clarified crude extracts containing medium and cells as described in Materials and Methods.
Interestingly, a distinct norvaline peak was detected in HPLC chromatograms of the anaerobic culture which was not found in aerobic control cultures (see Fig. 2). Norvaline increased significantly and accumulated to about 1 mM at 7 hours after the stirrer downshift. This was the highest concentration of a branched chain amino acid detected during the cultivation with the stirrer downshift. In contrast, the norvaline remained below the detection limit in all samples from the aerobic control fermentation (see Fig. 4).
Next, it was interesting to investigate whether norvaline accumulation occurs simply due to the accumulation of pyruvate, or whether additionally a deregulation of the synthesis of enzymes of the branched chain amino acid pathway is needed. Therefore, we studied the transcriptional profile of a number of marker genes during the shift to anaerobiosis (Figs. 5 and 6). The transcriptional profiles showed a clear induction of anaerobic metabolism by the stirrer downshift. The most significant event was the induction of pyruvate formate lyase (pflB), which responded immediately after the DOT dropped to zero, and interestingly even responded very sensitively to small changes in the process. For example, the pflB mRNA level immediately decreased when the culture run into glucose starvation for a short time at about 15 min after the anaerobic shift and it increased immediately after the DOT decreased to zero again (Fig. 5). Also the expression of lactate dehydrogenase (ldhA) behaved similarly. Also DNA-microarray analysis indicated a significant induction of the genes of anaerobic metabolism, such as the FNR regulon and the operons of formate dehydrogenase (fdh) and formate hydrogen lyase (hyc) which are included in formate degradation, already 20 min after the down-shift (Fig. 6). It should be remarked, however, that the pflB signal from the microarrays was different. In the DNA microarrays no induction of pflB was detected, which is remarkable as it contradicts the results with the sandwich hybridization assay and the finding that formate accumulated significantly.
A typical anaerobic reaction was the strong repression of the aceE and aceF mRNAs encoding polypeptides of the pyruvate dehydrogenase complex as observed by microarray analysis (not shown, see additional material). All other genes of the glycolysis were only marginally downregulated. Also the genes of the tricarbonic acid cycle reacted in a typical way [27, 28] (see Fig. 6), the mRNAs of the suc and sdh operons decreased very strongly. A significant decrease was also observed for the icdA mRNA encoding for isocitrate dehydrogenase, mdh mRNA encoding for malate dehydrogenase, and fumA encoding for the aerobic fumarate hydratase. A strong increase was seen for the mRNAs of enzymes of the left branch of TCA responsible for the reactions from oxaloacetate to succinate, such as fumB encoding the anaerobic fumarate hydratase and the frd operon encoding subunits of the fumarate reductase. The mRNA of citrate synthase (gltA) was slightly repressed and recovered later according to the DNA array data (Fig. 6a) and similar also in the sandwich hybridization (Fig. 5), although here the gltA level decreased below the detection limit, but showed a high expression in the last sample.
The data for the mRNAs of the branched chain amino acid pathway from the DNA arrays showed generally a decrease of all of the mRNAs after the oxygen downshift with a fast initial response between the samples collected 20 min before the oxygen downshift and 10 min after it (Fig. 6e). A more detailed dynamic response was obtained by analysis of some representative mRNAs from the different operons by sandwich hybridization. Interestingly, but not unexpected, this showed a fast and transient accumulation of the ilvA mRNA after the anaerobic shift. Although the increase was only approximately two-fold, the curve showed a similar shape as for pflB (Fig. 5). All other mRNAs of the pathway were not observed to react so clearly and fast to oxygen depletion. However, there was a second slower continuous increase of the mRNA levels of ilvA, leuA, and ilvG after 1.5 hours. In contrast, there was no significant derepression observed for the level of the ilvB and ilvI mRNAs coding of the large subunits of acetohydroxy acid synthetases (AHAS) I and III respectively.
These data suggest that it is unlikely that the synthesis of norvaline is due to an extra derepression of the leucine operon, but that the existing enzymes account for the synthesis. This was further confirmed by 2-dimensional electrophoresis of the proteome which indicated no significant increase of any of the enzymes of the branched chain amino acid synthetic pathway after the anaerobic shift (data not shown).
Discussion
The results presented here show that E. coli W3110 is able to produce the amino acid analogue norvaline after a shift from aerobic to anaerobic conditions in a culture on pure glucose mineral salt medium. This is new and interesting, as so far norvaline accumulation has been described only for mutant strains, e.g. for mutants of Serratia marcescens which are used for isoleucine production [5, 7], and for recombinant E. coli strains which express of recombinant proteins with a high content of leucine and which consequently decrease the cellular leucine pool [16]. It is surprising that norvaline is produced by a normal environmental shift. The conditions which have been shown here to result in norvaline accumulation are relevant when shake flask cultures run into oxygen limitation. Also large- scale processes which are characterized by imperfect mixing may result in norvaline accumulation, as it is known that high glucose concentration and oxygen limitation characterize feeding zones of large-scale bioreactors [29]. Although there is no specific cellular function known for norvaline in E. coli, it has been shown that norvaline can be incorporated in recombinant proteins instead of leucine, which is generally not wanted.
Pyruvate is a starting metabolite for the branched chain amino acid pathway and the synthesis of the amino acid analog norvaline from α-ketobutyrate. There is strong evidence that the synthesis of norvaline or norleucine is due to direct chain elongation of pyruvate over 2-ketobutyrate and 2-ketocaproate (for norleucine) if the derepression of the leucine operon is caused by depletion of the amino acid leucine through strong induction of recombinant proteins with an over-average content of leucine [15, 16]. In the case of our experimental set-up the leucine concentration was low due to cultivation on mineral salt medium and correspondingly the leucine operon was derepressed as is seen by the level of α-IPMS mRNA confirming the results by Bogosian et al. [15].
The critical factor in our experiments was the accumulation of pyruvate, caused by down-shifts of the oxygen level. Pyruvate accumulation and norvaline incorporation into a recombinant product has been observed by Apostol et al. [16] in aerobic conditions. Although the authors did not discuss the origin of this increase, it seems that the high pyruvate concentration in their cultivation resulted from changes in metabolic fluxes through the glycolysis due to expression of their recombinant product. Uncoupling of the glycolysis and respiration after strong induction of recombinant product have been observed earlier [30], but have not been attributed to fluxes in the amino acid pathways.
As the high pyruvate concentration in crude broth lysates and a derepression of the leucine operon were fulfilled in our experiments, we believe that norvaline accumulation occurred by chain elongation of pyruvate. Although a detailed metabolic flux analysis is needed for validation, this hypothesis is strongly supported by the high concentration of pyruvate, which is two orders of magnitude above the concentration of threonine and significantly above the apparent Km of the α-isopropylmalate synthase for pyruvate (10 mM, [24]). The direct chain elongation by the enzymes of the leuABCD operon is more likely than assuming the flux going over the long oxaloacetate and threonine pathway.
It may be remarked, that we also found an accumulation of aspartate and principally there should be the option that α-ketobutyrate is synthesized over the aspartate-threonine pathway. Interesting in this view is the anaerobic positive regulation of ilvA. IlvA mRNA was clearly shown to accumulate by measuring the dynamic response by sandwich hybridization although this response was not seen in the DNA array analysis, which only covered the first hour after the anaerobic shift with 3 samples collected in 30 min time space. However, despite the derepression of gene expression during anaerobic growth as monitored on the mRNA level (Fig. 5) and by protein synthesis (35S-methionine labeling of proteome and 2D electrophoresis, not shown) the amount of threonine deaminase was not significantly increasing (2D electrophoresis, not shown). Also, the level of threonine did not increase, which might be eventually expected if the flux through this pathway should be high. Therefore, we conclude that norvaline is produced by direct chain elongation of pyruvate as described before in recombinant processes.
Norvaline accumulation was only observed when we shifted the cells from aerobic to anaerobic conditions abruptly at a high glucose concentration. No norvaline accumulation was found when the aeration was shut off in a glucose limited chemostat and the glucose concentration increased gradually (data not shown), or when glucose limited cells in a chemostat growing with a specific growth rate of 0.1 h-1 obtained a glucose pulse. Similar chemostat experiments performed by Chassagnole et al. [31] showed that the cellular pyruvate level only increased to about 4 mM, which is far below the level which was e.g. detected by [16] and in our studies. Long term cultivation under lower growth rate decreases the glucose uptake capacity [32] and although experimental data are not available to our knowledge, it could be proposed that the lower influx of glucose in a culture growing with a low specific growth rate may negatively affect the accumulation of pyruvate. Therefore we see the chemostat data in agreement with the assumption that a high glucose flux into the glycolysis is an important parameter.
Finally, we want to highlight the fact that the accumulation of norvaline might be favored in E. coli K-12 strains by the frameshift mutation in the ilvG gene, an interesting point, which is currently under investigation.
Conclusion
In this study we have shown that norvaline can accumulate in the well-known E. coli K-12 strain W3110 if it is grown in mineral salt medium under conditions where fast growing cells undergo a shift to anaerobiosis. The important parameters seem to be a high expression of the enzymes of the leucine operon and the accumulation of pyruvate. It is likely that pyruvate is the substrate for direct keto chain elongation to α-ketobutyrate and further to α-ketovalerate from which norvaline is synthesized by transamination. The level of accumulated norvaline is very high. It remains to be investigated whether this might have relevance for the cell physiology. So far in most proteins where norvaline was incorporated the activity of the protein was maintained. The new mechanism of norvaline might be of direct biotechnological relevance if one aims to incorporate norvaline into proteins, e.g. for structural studies.
Our finding also may be of practical relevance for securing the production of non-modified recombinant proteins. The oxygen level e.g. in a shake flask culture or the occurrence of oxygen starvation zones in a large-scale bioreactor may have an impact on the appearance and incorporation of norvaline.
Methods
Strain and cultivation conditions
The strain used in this study was E. coli W3110 [F-IN(rrnD-rrnE)1].
The cultivations were performed in a Biostat C 15 L bioreactor with the DCU-3 controlling unit and MFCS-win supervisory system (Sartorius) with an initial working volume of 8 L. The mineral salt medium contained per liter: 14.6 g K2HPO4, 3.6 g NaH2PO4 × 2 H2O, 2.0 g Na2SO4, 2.47 g (NH4)2SO4, 0.5 g NH4Cl, 1.0 g (NH4)2-H-citrate, 2 mM MgSO4, 0.1 g thiamine hydrochloride, 0.1 mL antifoam 204 (Sigma) and 2 mL trace element solution [33]. The initial glucose concentration was 40 g L-1. The feed solution contained 650 g L-1 glucose. 2 mL L-1 of sterile filtered 1 M MgSO4 were added regularly per OD600 = 10 increase.
Two precultures were performed in LB medium and mineral salt medium with 10 g L-1 of glucose without antifoam agent consecutively at 37°C at a rotary shaker at 180 rpm. Main cultivations were started as batch cultures at a temperature of 37°C. The pH was kept at 7.0 by controlled addition of 25% ammonia solution. At the end of the exponential growth phase (cell dry weight about 16 g L-1) the stirrer rate was lowered from 1000 rpm to 500 rpm, to provoke oxygen limitation by decreased oxygen transfer. Constant glucose feed of 100 g L-1 h-1 was started 15 min after the oxygen drop which was enough to ensure glucose excess during the whole cultivation.
Analysis of cell growth
Cell growth was monitored by measurement of the absorbance (OD600) and cell dry weight as described earlier [34]. One unit of OD600 corresponds to a dry cell weight of 0.44 g L-1.
Amino acid analysis
The amino acids were analyzed from clarified crude broth lysates. Therefore broth samples, containing medium and cells, were taken and immediately shock-frozen in liquid nitrogen and stored at -20°C until the sample preparation. Samples, diluted to same cell density with 0.9% NaCl solution, were sonicated on ice for 2 × 30 sec with 30 sec cooling break between the sonication steps. After sonication the cell debris was removed by centrifugation for 10 min (+4°C, 16,100 × g) and the supernatant was purified from macromolecules by centrifugation for 60 min at 14,000 × g and +4°C using Microcon 3 kDa cut-off membranes (Millipore). The filtrate was diluted 20 times with 0.1 M HCl and OPA – precolumn derivatisation was used for the amino acid analysis. OPA reagent (Agilent) was diluted 5 times in 0.4 M borate buffer (Agilent) and equal amounts of diluted sample and OPA reagent were mixed for reaction. After approximately 1 min 20 μL of derivatisation reaction solution was injected into the Zorbax Eclipse column (Agilent). The inorganic eluent contained 0.03 M sodium acetate, 0.25% tetrahydrofurane (v/v) and 100 ppm sodium azide, and the pH was set to 7.2 with 1% acetic acid [v/v]. The organic eluent consisted of 80% acetonitrile and 20% of 0.03 M sodium acetate [v/v]. The HPLC equipment consisted of pump series (P580), autosampler (ASI-100), column oven (HTS-585) and fluorescence detector (RF-2000) from Dionex.
Analysis of metabolites and glucose
Acetate, pyruvate, formate, succinate, lactate, ethanol, and glucose were analyzed from medium samples, which were immediately centrifuged for 3 min at 16,100× g and +4°C. The supernatant was filtered (0.2 μm) and frozen in liquid nitrogen. Pyruvate was analyzed from clarified crude extracts which were prepared as described for amino acid analysis. The metabolites were analyzed in a Merck-Hitachi HPLC system (Model D-7000) with an ICSep COREGEL 87H3 column. An L-4259 UV-VIS detector (Merck-Hitachi) at 210 nm was used for the organic acids and a differential refractometer RI-71 (Merck) for glucose and ethanol.
mRNA analysis by sandwich hybridization
Samples for mRNA analysis were collected as described earlier [34]. Samples were shortly mixed by vortexing, divided in 0.5 mL aliquots and centrifuged for 3 min at 16,100 × g and +4°C. The pellets were resuspended in 250 μL of RNALater (Ambion, USA) and stored at -20°C until analysis. RNA was extracted using the total RNA kit (A&A Biotechnology, Poland) following the instructions of the kit. A sandwich hybridisation assay was performed as described earlier [35]. In vitro transcribed RNA molecules were used as standards for quantification of mRNA molecules in the samples (see table 1 for the primers used). The amplification of in vitro RNA was done as described earlier (28). For each gene of interest a detection probe labeled with digoxigenin (Roche, USA), a capture probe labeled by biotin (by probe manufacturer), and two unlabelled helper probes were designed. Different sets of probes were tested and the set producing the highest signal in the sandwich assay was chosen (see Table 1 for details of the probes).
Microarray analysis
Experimental procedures for GeneChip (Affymetrix, Santa Clara, CA, USA) were performed according to the Affymetrix GeneChip Expression Analysis Technical Manual. Affymetrix E. coli Genome 2.0 array was used. Sample preparation was equal to mRNA analysis by sandwich hybridization described in previous chapter.
The microarray data have been deposited in NCBI's Gene Expression Omnibus [36] and are accessible through GEO Series accession number GSE12006 http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE12006.
References
Nandi P, SEN GP: An antifungal substance from a strain of Bacillus subtilis. Nature. 1953, 172: 871-872. 10.1038/172871b0.
Kisumi M, Sugiura M, Chibata I: Biosynthesis of norvaline, norleucine, and homoisoleucine in Serratia marcescens. J Biochem. 1976, 80 (2): 333-339.
Kisumi M, Sugiura M, Takagi T, Chibata I: Norvaline accumulation by regulatory mutants of Serratia marcescens. J Antibiot (Tokyo). 1977, 30: 111-117.
Kisumi M, Sugiura M, Chibata I: Norleucine accumulation by a norleucine-resistant mutant of Serratia marcescens. Appl Environ Microbiol. 1977, 34: 135-138.
Sugiura M, Kisumi M, Chibata I: Biosynthetic pathway of beta-methylnorleucine, an antimetabolite produced by Serratia marcescens. J Antibiot (Tokyo). 1981, 34: 1283-1289.
Sugiura M, Kisumi M, Chibata I: beta-methylnorleucine, an antimetabolite produced by Serratia marcescens. J Antibiot (Tokyo). 1981, 34: 1278-1282.
Kisumi M, Sugiura M, Kato J, Chibata I: L-Norvaline and L-homoisoleucine formation by Serratia marcescens. J Biochem. 1976, 79 (5): 1021-1028.
Muramatsu R, Miura A, Misawa S: [A finding of isoleucine derivative in recombinant protein]. Tanpakushitsu Kakusan Koso. 2002, 47: 1299-1304.
Muramatsu R, Negishi T, Mimoto T, Miura A, Misawa S, Hayashi H: Existence of beta-methylnorleucine in recombinant hirudin produced by Escherichia coli. J Biotechnol. 2002, 93: 131-142. 10.1016/S0168-1656(01)00396-0.
Muramatsu R, Misawa S, Hayashi H: Finding of an isoleucine derivative of a recombinant protein for pharmaceutical use. J Pharm Biomed Anal. 2003, 31: 979-987. 10.1016/S0731-7085(02)00703-3.
Sunasara KM, Cramer SM, Hauer CR, Rupp RG, Shoup VA: Characterization of recombinant human brain-derived neurotrophic factor variants. Arch Biochem Biophys. 1999, 372: 248-260. 10.1006/abbi.1999.1501.
Lu HS, Tsai LB, Kenney WC, Lai PH: Identification of unusual replacement of methionine by norleucine in recombinant interleukin-2 produced by E. coli. Biochem Biophys Res Commun. 1988, 156: 807-813. 10.1016/S0006-291X(88)80916-1.
Tsai LB, Lu HS, Kenney WC, Curless CC, Klein ML, Lai PH, Fenton DM, Altrock BW, Mann MB: Control of misincorporation of de novo synthesized norleucine into recombinant interleukin-2 in E. coli. Biochem Biophys Res Commun. 1988, 156: 733-739. 10.1016/S0006-291X(88)80904-5.
Fenton D, Lai PH, Lu H, Mann M, Tsai L: Control of norleucine incorporation into recombinant proteins. U.S. patent no. 5, 599, 690. 1994
Bogosian G, Violand BN, Dorward-King EJ, Workman WE, Jung PE, Kane JF: Biosynthesis and incorporation into protein of norleucine by Escherichia coli. J Biol Chem. 1989, 264: 531-539.
Apostol I, Levine J, Lippincott J, Leach J, Hess E, Glascock CB, Weickert MJ, Blackmore R: Incorporation of norvaline at leucine positions in recombinant human hemoglobin expressed in Escherichia coli. J Biol Chem. 1997, 272: 28980-28988. 10.1074/jbc.272.46.28980.
Barker DG, Bruton CJ: The fate of norleucine as a replacement for methionine in protein synthesis. J Mol Biol. 1979, 133: 217-231. 10.1016/0022-2836(79)90531-X.
Budisa N, Steipe B, Demange P, Eckerskorn C, Kellermann J, Huber R: High-level biosynthetic substitution of methionine in proteins by its analogs 2-aminohexanoic acid, selenomethionine, telluromethionine and ethionine in Escherichia coli. Eur J Biochem. 1995, 230: 788-796. 10.1111/j.1432-1033.1995.tb20622.x.
Budisa N, Huber R, Golbik R, Minks C, Weyher E, Moroder L: Atomic mutations in annexin V – thermodynamic studies of isomorphous protein variants. Eur J Biochem. 1998, 253: 1-9. 10.1046/j.1432-1327.1998.2530001.x.
Gilles AM, Marliere P, Rose T, Sarfati R, Longin R, Meier A, Fermandjian S, Monnot M, Cohen GN, Barzu O: Conservative replacement of methionine by norleucine in Escherichia coli adenylate kinase. J Biol Chem. 1988, 263: 8204-8209.
Randhawa ZI, Witkowska HE, Cone J, Wilkins JA, Hughes P, Yamanishi K, Yasuda S, Masui Y, Arthur P, Kletke C: Incorporation of norleucine at methionine positions in recombinant human macrophage colony stimulating factor (M-CSF, 4–153) expressed in Escherichia coli : structural analysis. Biochemistry. 1994, 33: 4352-4362. 10.1021/bi00180a032.
Semmes OJ, Riehm JP, Rao KR: Substitution of norleucine for methionine residues in a crustacean pigment-dispersing hormone. Peptides. 1985, 6: 491-494. 10.1016/0196-9781(85)90115-9.
Violand BN, Siegel NR, Bogosian G, Workman WE, Kane JF: Techniques in Protein Chemistry. Edited by: Hugli TE. 1989, 315-326. San Diego: Academic Press
Kohlhaw G, Leary TR, Umbarger HE: Alpha-isopropylmalate synthase from Salmonella typhimurium . Purification and properties. J Biol Chem. 1969, 244: 2218-2225.
Bridger WA, Paranchych W: relA Gene control of bacterial glycogen synthesis. Can J Biochem. 1978, 56: 403-406.
Hecker M, Schroeter A: [3H-thymidine incorporation following isoleucine limitation in stringent and relaxed controlled strains of Escherichia coli]. Z Allg Mikrobiol. 1984, 24: 57-60. 10.1002/jobm.3630240111.
Vemuri GN, Eiteman MA, Altman E: Effects of growth mode and pyruvate carboxylase on succinic acid production by metabolically engineered strains of Escherichia coli. Appl Environ Microbiol. 2002, 68: 1715-1727. 10.1128/AEM.68.4.1715-1727.2002.
Thomas AD, Doelle HW, Westwood AW, Gordon GL: Effect of oxygen on several enzymes involved in the aerobic and anaerobic utilization of glucose in Escherichia coli. J Bacteriol. 1972, 112: 1099-1105.
Enfors SO, Jahic M, Rozkov A, Xu B, Hecker M, Jürgen B, Krüger E, Schweder T, Hamer G, O'Beirne D, Noisommit-Rizzi N, Reuss M, Boone L, Hewitt C, McFarlane C, Nienow A, Kovacs T, Trägårdh C, Fuchs L, Revstedt J, Friberg PC, Hjertager B, Blomsten G, Skogman H, Hjort S, Hoeks F, Lin HY, Neubauer P, van der LR, Luyben K, Vrabel P, Manelius Å: Physiological responses to mixing in large scale bioreactors. J Biotechnol. 2001, 85: 175-185. 10.1016/S0168-1656(00)00365-5.
Neubauer P, Lin HY, Mathiszik B: Metabolic load of recombinant protein production: Inhibition of cellular capacities for glucose uptake and respiration after induction of a heterologous gene in Escherichia coli. Biotechnol Bioeng. 2003, 83: 53-64. 10.1002/bit.10645.
Chassagnole C, Noisommit-Rizzi N, Schmid JW, Mauch K, Reuss M: Dynamic modeling of the central carbon metabolism of Escherichia coli. Biotechnol Bioeng. 2002, 79: 53-73. 10.1002/bit.10288.
Lin HY, Mathiszik B, Xu B, Enfors SO, Neubauer P: Determination of the maximum specific uptake capacities for glucose and oxygen in glucose-limited fed-batch cultivations of Escherichia coli. Biotechnol Bioeng. 2001, 73: 347-357. 10.1002/bit.1068.
Holme T, Arvidson S, Lindholm B, Pavlu B: Enzymes: laboratory-scale production. Process Biochem. 1970, 5: 62-66.
Neubauer A, Soini J, Bollok M, Zenker M, Sandqvist J, Myllyharju J, Neubauer P: Fermentation process for tetrameric human collagen prolyl 4-hydroxylase in Escherichia coli : improvement by gene optimisation of the PDI/beta subunit and repeated addition of the inducer anhydrotetracycline. J Biotechnol. 2007, 128: 308-321. 10.1016/j.jbiotec.2006.10.017.
Rautio J, Barken KB, Lahdenperä J, Breitenstein A, Molin S, Neubauer P: Sandwich hybridisation assay for quantitative detection of yeast RNAs in crude cell lysates. Microb Cell Fact. 2003, 2: 4- 10.1186/1475-2859-2-4.
Edgar R, Domrachev M, Lash AE: Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30: 207-210. 10.1093/nar/30.1.207.
Acknowledgements
Support by the European Structural Funds (Bioworld I and HTS-Bio projects) is kindly acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JS carried out the experiments and drafted the manuscript. CF participated in the experiments with the anaerobic shift simulator and performed part of the SH mRNA analyses. CL and JB performed the 2D electrophoresis experiments. JV performed the DNA arrays. PN conceived of the study, and participated in its design, coordination, and drafting of the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Soini, J., Falschlehner, C., Liedert, C. et al. Norvaline is accumulated after a down-shift of oxygen in Escherichia coli W3110. Microb Cell Fact 7, 30 (2008). https://doi.org/10.1186/1475-2859-7-30
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2859-7-30