Abstract
Background
An ameloblastoma is a benign odontogenic neoplasm with aggressive behaviour and high recurrence rates. The increased expression of matrix metalloproteinases (MMPs) has been reported in ameloblastomas. In the present study, we hypothesised that epigenetic alterations may regulate MMP expression in ameloblastomas.
Methods
We investigated the methylation status of the genes MMP-2 and MMP-9 in addition to mRNA transcription and protein expression in ameloblastomas. Methylation analysis was performed by both methylation-specific polymerase chain reaction (MSP-PCR) and restriction enzyme digestion to evaluate the methylation profile of MMP-2 and MMP-9 in 12 ameloblastoma samples and 12 healthy gingiva fragments, which were included as controls. Furthermore, we investigated the transcription levels of the genes by quantitative reverse-transcription PCR (qRT-PCR). Zymography was performed to verify protein expression in ameloblastomas.
Results
The ameloblastomas showed a high frequency of unmethylated MMP-2 and MMP-9, whereas the healthy gingival samples presented a sharp prevalence of methylated MMPs. Higher expression levels of MMP-9 were found in ameloblastomas compared to healthy gingiva. However, no significant differences in the MMP-2 mRNA expression between groups was found. All ameloblastomas showed positive expression of MMP-2 and MMP-9 proteins.
Conclusions
Our findings suggest that expression of MMP-9 is increased in ameloblastomas and is possibly modulated by unmethylation of the gene.
Similar content being viewed by others
Background
An ameloblastoma is a benign odontogenic tumour that exhibits a high recurrence risk, aggressive behaviour and local invasiveness[1]. Histologically, an ameloblastoma consists of epithelial strands or islands of ameloblastic epithelium. The peripheral cells are columnar, while the cells lying more centrally are fusiform to polyhedral and are loosely connected to each other[1].
Different studies have demonstrated genetic alterations in odontogenic tumours[2–4], but few studies have analysed epigenetic events in these tumours[5–7]. Methylation is an epigenetic alteration that plays an important role in controlling gene activity, embryonic development, and genomic imprinting. It has been associated with gene silencing by transcriptional inactivation[8]. DNA methylation or hypomethylation of the p16 p21 and LINE-1 genes was reported in ameloblastomas by our group and others[5, 9, 10], but the significance of this data remains to be determined.
Matrix metalloproteinases (MMPs) are zinc-dependent enzymes that are important in extracellular matrix remodelling and are associated with tumour growth and invasion through collagen matrix degradation[11]. The invasive characteristic of ameloblastomas has been associated with the expression of genes related to bone turnover and extracellular matrix remodelling; these include BMP RANKL and its receptor, MMP and TIMP[12–17]. As MMPs may be regulated by DNA methylation in malignant neoplasms[18, 19], such phenomenon might be important in ameloblastoma pathogenesis and should be investigated. Therefore, the purpose of this study was to investigate the association between MMP-2 and MMP-9 methylation and their mRNA transcription and protein expression in ameloblastomas.
Methods
Patients and tissue samples
Twelve fresh ameloblastoma specimens were collected during surgical care in the Department of Oral Surgery and Pathology, Universidade Federal de Minas Gerais, Brazil. These samples comprised eleven solid-multicystic follicular ameloblastomas and one unicystic case. Diagnoses were confirmed by histopathologic analysis based on the World Health Organization classification of histological typing of odontogenic tumours[1]. Other clinical data are shown in Table1. Twelve fragments of healthy gingival samples with no clinical evidence of inflammation were collected during third molar extractions and used as controls. The samples were obtained following informed consent and with the approval of the Universidade Federal de Minas Gerais Ethics Committee (reference number 266/11).
DNA isolation and methylation analysis of MMP-2 and MMP-9
Genomic DNA was isolated from the tissue samples using a Qiagen DNeasy Tissue Kit (Qiagen Inc., Valencia, CA, USA) according to the manufacturer’s instructions. MethPrimer software[20] was used to search CpG islands and sparse CG dinucleotides. Distinct methods are suggested to analyse methylation profiles according to the presence of CpG islands or sparse CG dinucleotides located in the promoter region or in exons near to that region[21].
To assess the MMP-2 gene CpG island methylation, genomic DNA was modified by sodium bisulfite as described previously[6] and subsequently amplified with primer sets designed to specifically recognise methylated (F 5’-GCGGTTATACGTATCGAGTTAGC-3’ and R 5’-ACTCTTTATCCGTTTTAAAAACGAC-3’; 205 bp) and unmethylated DNA (F 5’-GGTGGTTATATGTATTGAGTTAGTGA-3’ and R 5’-ACTCTTTATCCATTTTAAAAACAAC-3’ 206 bp). Bisulfite-treated unmethylated DNA from (peripheral blood mononuclear cells) cells was used as a positive control for unmethylated amplification of the MMP-2 gene. Methylation-induced DNA of same cells by the MSssI methylase enzyme (New England Biolabs, Beverly, USA) was used as positive control for methylated amplification.
The methylation-sensitive restriction enzymes HhaI and AciI (New England BioLabs, Beverly, MA, USA) were used to assess the methylation of CG dinucleotides in the MMP-9 promoter, including the CG sites located at positions -35, -185, -223, -233, as described previously[21]. Restriction enzymes cleave DNA at unmethylated CG sites, but they are unable to cut methylated cytosines. Analysis using a bioinformatics web site (http://www.restrictionmapper.org) showed that the HhaI enzyme cleaves the restriction site at position -35 and that the other sites are cleaved by AciI. The CG dinucleotides analysed in this study are located close to the transcription start of the MMP-9 gene. Two hundred nanograms of genomic DNA was digested separately with each of the restriction enzymes HhaI and AciI according to manufacturer's protocol to cleave the specific regions containing CG sites (New England BioLabs, Beverly, MA, USA). Digestion was followed by PCR amplification (primers: F 5’-GCTTCATCCCCCTCCCTCC-3’, R 5’-AGCACCAGGACCAGGGGC-3’; 369 bp). PCR products were subjected to electrophoresis in 6.5% polyacrylamide gels. While methylated cytosine produces a band equivalent to that of control methylated DNA of placenta tissue, the cleavage by restriction enzyme at unmethylated CpG induces DNA strand breaks, and thus no band is detected. In each PCR reaction, undigested DNA of each sample was also carried out as controls. Undigested and digested PCR products were electrophoresed in parallel. Human unmethylated DNA (Qiagen Inc., Valencia, CA, USA), which is sensitive to action of the enzyme, was also used as unmethylated positive control.
RNA extraction and Quantitative Real-time PCR (qRT-PCR) of MMP-2 and MMP-9
Total RNA was extracted from tissue samples using Trizol reagent according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA, USA). RNA integrity was analysed by 1% agarose gel electrophoresis. Reverse transcription of 1 μg of RNA to cDNA was performed using SuperScript III First-Strand (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. Primer sequences were designed using the PrimerExpress software (v.3, Applied Biosystems) as follows: MMP-2 (F: 5’-AGCTCCCGGAAAAGATTGATG-3’; R: 5’-CAGGGTGCTGGCTGAGTAGAT-3’, 101 bp) and MMP-9 (F: 5’-GAGGTTCGACGTGAAGGCGCAGAT-3’; R: 5’-CATAGGTCACGTAGCCCACT TGGT3’, 200 bp). All reactions were run in duplicate in a StepOne Real time PCR System using the SYBR-green fluorescence quantification system (Applied Biosystems, Warrington, UK). The comparative C t method was used[22]. Expression levels of the MMP-2 and MMP-9 genes relative to a calibrator sample (placenta tissue) were obtained by normalisation to endogenous β-actin.
Gelatin zymography
Ameloblastoma protein was extracted and subjected to electrophoresis under nonreducing conditions on SDS-polyacrylamide gels copolymerised with 1 mg/ml gelatin (Sigma Chemical Co, St Louis, MO, USA) as previously described[23]. After electrophoresis, the gels were washed in 2.5% Triton-X 100 and incubated for at least 18 h at 37°C in incubation buffer (50 mM Tris–HCl, pH 7.5, containing 5 mM CaCl2, 100 mM NaCl, 0.01% Triton X-100). Zymographic gels were stained in 0.2% Coomassie Brilliant Blue R-250 and de-stained. The gels were scanned to analyse the bands representative of MMPs, according to molecular weight. Analysis of protein expression in healthy gingiva was not performed due to the scarcity of tissue samples.
Statistical analysis
Mann–Whitney tests were used to compare the relative quantification of MMP-2 and MMP-9 between groups. Chi-squared or Fisher’s exact were used when appropriate. The analyses were carried out using SPSS 17.0 software, and probability values <0.05 were considered statistically significant.
Results
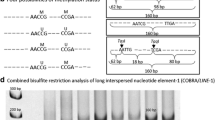
MMP-2 and MMP-9 methylation statuses are shown in Table2 and represented in Figure1. While all healthy gingival samples showed MMP-2 methylation, approximately half of ameloblastomas were unmethylated. Similarly, an increased frequency of unmethylated MMP-9 of specific CG sites digested by HhaI was identified in the ameloblastomas. Almost all of the ameloblastoma samples showed an unmethylated profile for MMP-9. No difference was found in the methylation of CG sites digested by Acil among the groups studied.
Representative figure of the methylation analysis. A: Methylation status of MMP-2 in ameloblastoma. M: PCR products when amplified by methylated primers (205 bp); U: PCR products when amplified by unmethylated primers (206 bp); +M: positive control for methylated reaction; +U: positive control for unmethylated reaction. -M and -U: negative controls without DNA. Lines 1 to 3 represent DNA from ameloblastoma samples. B: Methylation status of MMP-9 in ameloblastoma. DNA samples were digested by the AciI restriction enzyme followed by PCR, flanking the restriction sites. Absent band indicates unmethylated profile (U) due to DNA cleavage by the restriction enzyme. Presence of the PCR band represents methylated profile (M) of the MMP-9 gene. +M: methylated positive control; +U: unmethylated positive control; - : negative control without DNA.
The qRT-PCR results are summarised in Figures 2a and2b. Higher expression levels of MMP-9 were found in ameloblastomas compared to healthy gingiva. However, significant differences in the MMP-2 mRNA expression levels were not found.
When we investigated the influence of the methylation status of both genes on their transcription, no association was found between MMP-2 transcription and its methylation in ameloblastomas (p = 0.319). Almost all of the tumour samples showed an unmethylated MMP-9 pattern in conjunction with increased mRNA levels. As most of the ameloblastomas were unmethylated at the MMP-9 gene, considering all of the restriction sites, it was not possible to statistically compare the transcription of the gene in the cases with or without methylated sequences.
All of the ameloblastoma samples showed expression of MMP-2 and MMP-9 proteins, as verified by zymography (Figure3). However, pro-MMP-2 and pro-MMP-9 forms were not identified in ameloblastomas.
Discussion
The underlying molecular pathways associated with the pathogenesis of ameloblastomas are not well established yet. Previous investigations have assessed the molecular and genetic alterations related mainly to apoptosis, allelic loss of tumour suppressor genes, deregulation of the Sonic Hedgehog signalling pathway, and the clonality of these tumours[3, 24, 25].
Matrix metalloproteinases are involved in the degradation of collagen, as well as bone matrix, and have been shown to play a key role in the local invasiveness of ameloblastoma cells[15, 26]. Overexpression of MMP-2 and MMP-9 was associated with the infiltrative behaviour of ameloblastomas, as well as that of several malignant neoplasms[17, 27]. The suppression of MMP-2 activity was able to inhibit the invasiveness of ameloblastoma cells in vitro[14, 15]. Furthermore, it was suggested that increased expression of MMP-9 may be involved in the proliferation and invasive behaviour of ameloblastomas[26].
Some papers, including studies from our research group, have demonstrated epigenetic alterations in odontogenic tumours[5, 6, 9, 10, 28]. In the present study, we hypothesised that methylation may regulate the expression of MMP-2 and MMP-9 in ameloblastomas. We also investigated the association between methylation and the transcription levels of these genes. As most of the ameloblastoma samples were of the solid follicular type, we were not able to analyse possible associations between each clinical or histological type and the molecular data.
MMPs play an important role in collagen matrix remodelling in physiologic and pathologic processes, such as those found in basal membranes, dental follicle tissue and tumour metastasis[27, 29]. Although MMP-2 is related to ameloblastoma pathogenesis, it seems to be constitutively expressed in physiologic tissues and many cell types and to exhibit characteristics of a housekeeping gene[30–32]. Perhaps this could explain the similar expression levels of MMP-2 mRNA in ameloblastomas and healthy gingiva. Furthermore, our data suggest that MMP-2 expression in ameloblastomas may not be modulated by methylation because the methylation profile for this gene did not correlate with MMP-2 transcript levels in this odontogenic tumour.
The ameloblastomas presented an unmethylated profile of MMP-2 and MMP-9 genes compared to gingiva. Furthermore, along with unmethylated MMP-9, this tumour showed increased transcription of MMP-9 when compared to the control group. The important role of methylation in epigenetic silencing is well established, particularly through regulatory mechanisms of transcription. Accordingly, our data suggest that an unmethylated profile of the MMP-9 gene in ameloblastomas may be a permissive event allowing the binding of transcription factors to DNA, thus favouring MMP-9 gene transcription.
All of the ameloblastomas showed MMP-9 protein expression and were mostly unmethylated for MMP-9, so it was not possible to assess if the transcription of the gene was correlated with its methylation status. However, our study suggests that the increased transcription of MMP-9 in ameloblastomas could possibly be influenced by unmethylation of the gene. The evident protein expression, identified by zymography, provides additional evidence supporting the possible gene regulation by unmethylated MMP-9. It is interesting to note that hypomethylation of the MMP-2 and MMP-9 genes increases gene expression and contributes to cancer cell invasiveness and tumourigenesis in malignant neoplasms, such as prostate cancer and lymphoma[18, 33].
Conclusion
In conclusion, our study provides new insights into the epigenetic regulation of MMPs in ameloblastomas and points to the hypomethylation of MMP-9 as a possible mechanism involved in the increased transcription of the gene in this tumour. However, functional studies are needed to better explain the role the methylation of matrix metalloproteinases plays in the pathogenesis of ameloblastoma.
References
Barnes L, Eveson JW, Reichart PA, Sidransky D: World Health Organization classification of tumors: pathology and genetics of tumours of the head and neck. 2005, Lyon: IARC, 296-300.
Barreto DC, Gomez RS, Bale AE, Boson WL, De Marco L: PTCH gene mutations in odontogenic keratocysts. J Dent Res. 2000, 79: 1418-1422. 10.1177/00220345000790061101.
Nodit L, Barnes L, Childers E, Finkelstein S, Swalsky P, Hunt J: Allelic loss of tumor suppressor genes in ameloblastic tumors. Mod Pathol. 2004, 17: 1062-1067. 10.1038/modpathol.3800147.
Kumamoto H: Molecular pathology of odontogenic tumors. J Oral Pathol Med. 2006, 35: 65-74. 10.1111/j.1600-0714.2006.00380.x.
Kitkumthorn N, Mutirangura A: LINE-1 methylation difference between ameloblastoma and keratocystic odontogenic tumor. Oral Dis. 2010, 16: 286-291. 10.1111/j.1601-0825.2009.01640.x.
Moreira PR, Guimarães MM, Guimarães AL, Diniz MG, Gomes CC, Brito JA, Gomez RS: Methylation of P16, P21, P27, RB1 and P53 genes in odontogenic keratocysts. J Oral Pathol Med. 2009, 38: 99-103.
Pan S, Dong Q, Sun LS, Li TJ: Mechanisms of inactivation of PTCH1 gene in nevoid basal cell carcinoma syndrome: modification of the two-hit hypothesis. Clin Cancer Res. 2010, 16: 442-450. 10.1158/1078-0432.CCR-09-2574.
Esteller M: CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002, 21: 5427-5440. 10.1038/sj.onc.1205600.
Zhong M, Yan Y, Wang J, Liu J, Li L, Gong YB: DNA methylation of human telomerase reverse transcriptase promoter region in human ameloblastoma. Zhonghua Kou Qiang Yi Xue Za Zhi. 2007, 42: 304-307.
Moreira PR, Guimarães MM, Gomes CC, Diniz MG, Brito JA, de Castro WH, Gomez RS: Methylation frequencies of cell-cycle associated genes in epithelial odontogenic tumours. Arch Oral Biol. 2009, 54: 893-897. 10.1016/j.archoralbio.2009.07.006.
Hadler-Olsen E, Fadnes B, Sylte I, Uhlin-Hansen L, Winberg JO: Regulation of matrix metalloproteinase activity in health and disease. FEBS J. 2011, 278: 28-45. 10.1111/j.1742-4658.2010.07920.x.
Sandra F, Hendarmin L, Kukita T, Nakao Y, Nakamura N, Nakamura S: Ameloblastoma induces osteoclastogenesis: a possible role of ameloblastoma in expanding in the bone. Oral Oncol. 2005, 41: 637-644. 10.1016/j.oraloncology.2005.02.008.
Kumamoto H, Ooya K: Expression of bone morphogenetic proteins and their associated molecules in ameloblastomas and adenomatoid odontogenic tumors. Oral Dis. 2006, 12: 163-170. 10.1111/j.1601-0825.2005.01177.x.
Wang A, Zhang B, Huang H, Zhang L, Zeng D, Tao Q, Wang J, Pan C: Suppression of local invasion of ameloblastoma by inhibition of matrix metalloproteinase-2 in vitro. BMC Cancer. 2008, 8: 182-10.1186/1471-2407-8-182.
Zhang B, Zhang J, Huang HZ, Chen WL, Tao Q, Zeng DL, Zhang LT, Xu JH: Inhibition of ameloblastoma invasion in vitro and in vivo by inhibitor of metalloproteinase-2 activity. J Oral Pathol Med. 2009, 38: 731-736. 10.1111/j.1600-0714.2009.00771.x.
Zhang B, Zhang J, Huang HZ, Xu ZY, Xie HL: Expression and role of metalloproteinase-2 and endogenous tissue regulator in ameloblastoma. J Oral Pathol Med. 2010, 39: 219-222. 10.1111/j.1600-0714.2009.00827.x.
Qian Y, Huang HZ: The role of RANKL and MMP-9 in the bone resorption caused by ameloblastoma. J Oral Pathol Med. 2010, 39: 592-598. 10.1111/j.1600-0714.2009.00882.x.
Shukeir N, Pakneshan P, Chen G, Szyf M, Rabbani SA: Alteration of the methylation status of tumor-promoting genes decreases prostate cancer cell invasiveness and tumorigenesis in vitro and in vivo. Cancer Res. 2006, 66: 9202-9210. 10.1158/0008-5472.CAN-06-1954.
Chernov AV, Sounni NE, Remacle AG, Strongin AY: Epigenetic control of the invasion-promoting MT1-MMP/MMP-2/TIMP-2 axis in cancer cells. J Biol Chem. 2009, 284: 12727-12734. 10.1074/jbc.M900273200.
Li LC, Dahiya R: MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002, 18: 1427-1431. 10.1093/bioinformatics/18.11.1427.
Roach HI, Yamada N, Cheung KS, Tilley S, Clarke NM, Oreffo RO, Kokubun S, Bronner F: Association between the abnormal expression of matrix-degrading enzymes by human osteoarthritic chondrocytes and demethylation of specific CpG sites in the promoter regions. Arthritis Rheum. 2005, 52: 3110-3124. 10.1002/art.21300.
Livak KJ, Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001, 25: 402-408. 10.1006/meth.2001.1262.
Snoek-van Beurden PA, Von den Hoff JW: Zymographic techniques for the analysis of matrix metalloproteinases and their inhibitors. Biotechniques. 2005, 38: 73-83. 10.2144/05381RV01.
Gomes CC, Duarte AP, Diniz MG, Gomez R: Current concepts of ameloblastoma pathogenesis. J Oral Pathol Med. 2010, 39: 585-591. 10.1111/j.1600-0714.2010.00908.x.
Farias LC, Gomes CC, Brito JA, Galvão CF, Diniz MG, de Castro WH, Bernardes VD, De Marco LA, Gomez RS: Loss of heterozygosity of the PTCH gene in ameloblastoma. Hum Pathol. 2012, 43: 1229-1233. 10.1016/j.humpath.2011.08.026.
Kumamoto H, Yamauchi K, Yoshida M, Ooya K: Immunohistochemical detection of matrix metalloproteinases (MMPs) and tissue inhibitors of metalloproteinases (TIMPs) in ameloblastomas. J Oral Pathol Med. 2003, 32: 114-120. 10.1034/j.1600-0714.2003.00086.x.
Zhou CX, Gao Y, Johnson NW, Gao J: Immunoexpression of matrix metalloproteinase-2 and matrix metalloproteinase-9 in the metastasis of squamous cell carcinoma of the human tongue. Aust Dent J. 2010, 55: 385-389. 10.1111/j.1834-7819.2010.01258.x.
Moreira PR, Cardoso FP, Brito JA, Batista AC, Gomes CC, Gomez RS: Hypomethylation of tumor suppressor genes in odontogenic myxoma. Braz Dent J. 2011, 22: 422-427. 10.1590/S0103-64402011000500013.
Omar NF, Gomes JR, Neves JD, Salmon CR, Novaes PD: MT1-MMP expression in the odontogenic region of rat incisors undergoing interrupted eruption. J Mol Histol. 2011, 42: 505-511. 10.1007/s10735-011-9356-0.
Huhtala P, Chow LT, Tryggvason K: Structure of the human type IV collagenase gene. J Biol Chem. 1990, 265: 11077-11082.
Crowther M, Goodall S, Jones JL, Bell PR, Thompson MM: Localization of matrix metalloproteinase-2 within the aneurysmal and normal aortic wall. Br J Surg. 2000, 87: 1391-1400. 10.1046/j.1365-2168.2000.01554.x.
Goodall S, Crowther M, Hemingway DM, Bell PR, Thompson MM: Ubiquitous elevation of matrix metalloproteinase-2 expression in the vasculature of patients with abdominal aneurysms. Circulation. 2001, 104: 304-309. 10.1161/01.CIR.104.3.304.
Chicoine E, Estève PO, Robledo O, Van Themsche C, Potworowski EF, St-Pierre Y: Evidence for the role of promoter methylation in the regulation of MMP-9 gene expression. Biochem Biophys Res Commun. 2002, 297: 765-772. 10.1016/S0006-291X(02)02283-0.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1472-6890/12/11/prepub
Acknowledgements
The authors gratefully acknowledge grants from Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and MS/SCTIE/Decit, Brazil. Professor RS Gomez is a research fellow of CNPq.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RSG, CCG, and LCF participated in the study design and drafted the manuscript. LCF and MCS performed the experiments. EFF performed the statistical analysis. WHC and JCTL collected the ameloblastoma samples and clinical information. RSG and CCG reviewed all of the histological diagnoses. All of the authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Farias, L.C., Gomes, C.C., Rodrigues, M.C. et al. Epigenetic regulation of matrix metalloproteinase expression in ameloblastoma. BMC Clin Pathol 12, 11 (2012). https://doi.org/10.1186/1472-6890-12-11
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1472-6890-12-11