Abstract
Background
Pediocin-like bacteriocins, ribosomally-synthesized antimicrobial peptides, are generally coexpressed with cognate immunity proteins in order to protect the bacteriocin-producer from its own bacteriocin. As a step for understanding the mode of action of immunity proteins, we determined the crystal structure of PedB, a pediocin-like immunity protein conferring immunity to pediocin PP-1.
Results
The 1.6 Å crystal structure of PedB reveals that PedB consists of an antiparallel four-helix bundle with a flexible C-terminal end. PedB shows structural similarity to an immunity protein against enterocin A (EntA-im) but some disparity to an immunity protein against carnobacteriocin B2 (ImB2) in both the C-terminal conformation and the local structure constructed by α3, α4, and their connecting loop. Structure-inspired mutational studies reveal that deletion of the last seven residues of the C-terminus of PedB almost abolished its immunity activity.
Conclusion
The fact that PedB, EntA-im, and ImB2 share a four-helix bundle structure strongly suggests the structural conservation of this motif in the pediocin-like immunity proteins. The significant difference in the core structure and the C-terminal conformation provides a structural basis for the classification of pediocin-like immunity proteins. Our mutational study using C-terminal-shortened PedBs and the investigation of primary sequence of the C-terminal region, propose that several polar or charged residues in the extreme C-terminus of PedB which is crucial for the immunity are involved in the specific recognition of pediocin PP-1.
Similar content being viewed by others
Background
Bacteriocins are ribosomally-synthesized antimicrobial peptides produced by bacteria. Most bacteriocins are generally synthesized as prepeptides and can be classified into several classes depending on their post-translational processing and the mode of action [1–3]. Type I bacteriocins, of which a nisin is most well-known, undergo post-translational modifications such as formation of dehydrated residues and lanthionine bridges [4]. In contrast, type IIa bacteriocins are matured by simple cleavage of a leader peptide and characterized by a conserved YGNGVXC motif in the N-terminus [5–7]. These bacteriocins contain 37~48 residues and show potent activity against related Gram-positive bacteria, e.g. Listeria spp. [8, 9]. NMR studies revealed that type IIa bacteriocins consists of two domains, a cationic N-terminal domain and a hydrophobic C-terminal domain [10–12]: the N-terminal domain interacts with the anionic cell surface of Gram-positive bacteria [13, 14], while the C-terminal domain participates in the membrane permeabilization [7, 15, 16].
In general, type IIa bacteriocins are coexpressed with cognate immunity proteins (pediocin-like immunity proteins) composed of 88~115 amino acids, in order to protect the organism from the antimicrobial activity of its own bacteriocin [17–21]. It is known that the immunity proteins usually exhibit high specificity against their cognate bacteriocins. In contrast to the type IIa bacteriocins that exhibit high sequence conservation in their N-terminus, the degree of sequence homology between the pediocin-like immunity proteins varies (5~85%), which provides the basis for classification of them into three subgroups (A, B, and C) [22, 23]. The mode of action of immunity proteins and the basis of their specificity is still unclear, although the C-terminal half of immunity proteins is known as an important determinant for the specific recognition of their cognate bacteriocins [24, 25]. However, because pediocin-like immunity proteins don't interact directly with their cognate bacteriocins [6, 20, 26] and exist in cytoplasm [18, 20], it has been proposed that they may act by interfering with the formation of a functional pore complex in the membrane or blocking the pore itself. The accumulated structural information for certain class of immunity proteins could generally provide an important clue to their mode of action. However, although a number of pediocin-like immunity proteins have been identified and characterized, so far, only two structures have been reported: immunity proteins conferring immunity to enterocin A (EntA-im) [27] which belongs to subgroup A, and carnobacteriocin B2 (ImB2) [28] which belongs to subgroup C.
Pediocin secreted by Pediococcus spp. is a representative of the type IIa bacteriocins. Among pediocins, pediocin PA-1 and its operon of Pediococcus acidilactici PAC1.0 is the most well known [26, 29]. Recently, we identified another pediocin (pediocin PP-1) operon (pedA, pedB, pedC, and pedD) from Pediococcus pentosaceus CBT-8, which is isolated from the Korean traditional fermentative food, kimchi. Based on the remarkable degree of sequence identity (~99%) of PedB of P. pentosaceus to that of P. acidilactici, we regarded PedB as the immunity protein against pediocin PP-1. PedB is a small positively charged protein with 112 amino acids and belongs to subgroup A. No transmembrane helices are predicted from sequence analysis. To provide a structural insight into the function of PedB, we determined the three dimensional structure of PedB at 1.6 Å resolution and suggested the basis for the structural classification of pediocin-like immunity proteins, and examined the role of the C-terminal end of PedB.
Results and discussion
PedB confers immunity to pediocin PP-1
We assumed PedB as an immunity protein to pediocin PP-1 on the basis of sequence identity to that of P. acidilactici (only one conservative substitution (Glu for Asp) at a position of 104 exists between two PedB proteins for pediocin PP-1 and pediocin PA-1) (Figure 1b). To investigate whether PedB confers immunity to pediocin PP-1, we introduced the pedB gene into the bacteriocin-sensitive Lactobacillus sakei NCFB 2714 strain [24] and tested the susceptibility against ammonium sulphate-precipitated fermentate of P. pentosaceus containing pediocin PP-1 (See Methods). Because the pedB gene doesn't have its own promoter, we used the promoter of sodA encoding Mn-containing superoxide dismutase in Bacillus subtilis [30, 31]. L. sakei harboring the pedB gene was about 26 times less susceptible compared to the strain possessing only the sodA promoter, indicating that PedB successfully acted as the immunity protein to pediocin PP-1 (Figures 1c &4). However, when PedB purified from E. coli was mixed with pediocin PP-l solution, PedB failed to inactivate the bacteriocin (see Additional file 1), which is in line with the previous report that pediocin-like immunity proteins don't directly interact with their cognate bacteriocins [6, 20, 26].
(A) Sequence alignment of pediocin PP-1 from P. pentosaceus (pediocinPP) with pediocin PA-1 from P. acidilactici (pediocinPA), EnterocinA, and Carnobacteriocin B2 (CarnobacB2). The substitution between pediocin PP-1 and pediocin PA-1 is indicated by black box. The color scheme of white on dark grey indicates the consensus residue derived from the occurrence of >70% of a single residue at a given position. (B) Structure-based alignment of PedB from P. pentosaceus (PedB-PP) with PedB from P. acidilactici (PedB-PA), EntA-im, and ImB2. Secondary structure of PedB is presented above the alignment. The additional fifth helix of ImB2 is indicated by black bar. The color scheme is same to (A). The conservative substitution between PedB proteins for pediocin PP-1 and pediocin PA-1 is indicated by black box. (C) Pediocin PP-1 susceptibility of L. sakei strain harboring pedB gene and control plasmid. The MIC is the concentration of bacteriocin that inhibited growth of the indicator strain by 50%. The immunity activity is presented as the -fold increase in MIC observed for strains expressing PedB variants relative to MICs for strains containing only the control plasmid. The results represent the averaged data from at least three experiments. The psodA indicates the promoter of sodA encoding Mn-containing superoxide dismutase in B. subtilis. (D) Determination of oligomeric state of PedB. Oligomeric state of PedB was analysed by gel filtration chromatography. Predicted molecular mass of PedB (See Methods) is 14,428 Da (Kav = 0.42), indicating that PedB exists as a monomer in solution.
Overall structure of PedB
The native PedB and selenomethionine-substituted PedB mutant (L24M) (See Methods) were crystallized under similar conditions. The initial model was built according to the phased electron density map obtained by using anomalous scattering differences of selenium atoms in the selenomethionyl L24M crystal. A final model for native PedB was refined subsequently to 1.6 Å resolution against native data. The native PedB crystals contain one molecule in the asymmetric unit. Out of the total 112 amino acids in native PedB, residues 7–93 are visible in the crystal structure. No significant structural difference was observed between the native and mutant PedB (r.m.s.d of 0.3 Å for all Cα atoms).
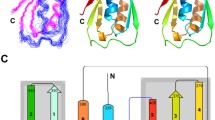
The crystal structure reveals that PedB forms a compact globular domain composed of four helices (Figure 2b). They are arranged so that α1 and α3 run in the same direction and α2 and α4 in the opposite direction, forming an antiparallel four-helix bundle (Figure 2b). The relative disposition of the four helices of PedB is maintained mainly by a network of hydrophobic interactions in the core of the molecule: Phe17, Leu20, and Leu24 from α1 (residues 9–26), Leu48 from α2 (residues 31–49), Val61 and Ile65 from α3 (residues 55–72), Leu74 and Phe76 from the loop between α3 and α4, Leu84 and Ile87 from α4 (residues 80–92) form a hydrophobic core (Figure 2c). This structure is further stabilized by several polar interactions between helices and between helix and loop: the side-chain of Gln21 makes polar contact with Glu41, Tyr35 with Gln68, Gln52 with Gln57, Tyr62 with Asn88, His27 with Tyr33 and Glu80 (figure not shown). The antiparallel helices are linked by a short loop of four to seven residues between them. The C-terminal end of PedB (residues 94–112) that is thought to be important for immunity has no electron density, suggesting that this region is flexible. We could not find any oligomeric contacts between PedB monomers in crystal packing, which is consistent with the result that PedB exists as a monomer in solution (Figure 1d).
(A) A 2Fo-Fc electron density map of the Tyr62 residue contoured at 1σ. The hole in the map indicates the high quality and resolution of the electron density map. (B) A ribbon diagram showing the overall structure (left) and the top view (right) of PedB shown with secondary structures labeled. (C) A ribbon diagram showing the hydrophobic core of PedB with secondary structures and important residues labled. (D) Surface charge representation of PedB on the view of α2 and α3. Blue and red colors indicate positive and negative charge, respectively.
Structural comparison of PedB to EntA-im and ImB2
Until now, two structures for pediocin-like immunity proteins, the crystal structure of EntA-im [27] and the solution structure of ImB2 [28], have been determined. Although PedB exhibits a limited sequence identity to EntA-im (14%) and ImB2 (6%) (Figure 1b), these three immunity proteins share an antiparallel left twisted four-helix bundle as a conserved scaffold. In addition, a positively charged region whose physiological function is obscure but which is commonly found in EntA-im, ImB2, and even in the structure models of pediocin-like immunity proteins [27, 28], is also observed in the surface representation of PedB. In PedB, the positively charged region is formed by residues from α2 (Lys46) and α3 (Arg56, Arg59 and Lys 64) (Figure 2d).
Pediocin-like immunity proteins are divided into three subgroups on the basis of the sequence homology [23, 28]. PedB and EntA-im belong to the subgroup A, and ImB2 belongs to the subgroup C. The fact that three immunity proteins share a common scaffold but are classified into different subgroups, aroused an interest in whether each subgroup has a conserved structural fingerprint. To test this, we compared the structure of PedB with those of EntA-im and ImB2, respectively. As expected, the structure of PedB is well-superimposed to that of EntA-im with r.m.s.d. of 1.3 Å for all Cα atoms (Figure 3b). They show a good agreement in the interhelical angles and length of helices. The structural similarity between PedB and EntA-im is much higher than expected from the degree of sequence identity [32].
(A) Ribbon diagrams of PedB (left), EntA-im (middle), and ImB2 (right) shown with secondary structures labeled. The axes of α3 and α4 are indicated by dotted lines. The angle between α3 and α4 is 33° for PedB and 31° for EntA-im. (B) Stereo view of the superimposed structures of PedB (red) and EntA-im (cyan). The flexible C-terminal loop of EntA-im that is thought to be important for its immunity is highlighted by a black ellipse. N and C indicate the N- and C-terminus of PedB, respectively.
(A) A graphic display of the immunity activity of L. sakei strains possessing PedB and PedB variants to pediocin PP-1. The results are presented using the same methods as in Figure 1c. The results represent the averaged data from at least three experiments, and the standard deviation values are indicated in each bar. (B) Far-UV CD spectra of PedB and C-terminal shortened PedB variants. CD spectra (190~250 nm) of 25 μg/ml ml wild type PedB and PedB variants were obtained to compare the secondary structure at 25°C. (C) Near-UV CD spectra of PedB and C-terminal shortened PedB variants. CD spectra (240~360 nm) of 200 μg/ml wild type PedB and PedB variants were obtained to compare the tertiary structure and protein folding at 25°C.
In contrast, compared with PedB and EntA-im, ImB2 shows a considerable disparity in both the C-terminal end and the core structure. Consistently, the r.m.s.d. value of 2.8 Å between PedB and ImB2 is higher than that of 1.3 Å between PedB and EntA-im. ImB2 contains an additional helix at the C-terminus, whereas the C-termini of PedB and EntA-im are flexible. In addition, the starting region (residues 31–33) of the second helix of PedB and EntA-im forms a 310 helix, which is not found in ImB2 (Figures 3a). The most remarkable structural difference in the four-helix bundle scaffold between PedB/EntA-im (subgroup A) and ImB2 (subgroup C) is observed in the local structure constructed by α3, α4, and a connecting loop. In PedB and EntA-im, α3, α4, and a straightened loop between them form a triangle-like conformation with an interhelical angle of ~30° (Figure 3a). Two hydrophobic residues from the connecting loop (Leu74 and Phe76 of PedB, and Ile67 and Phe69 of EntA-im) penetrate into the interhelical space between α3 and α4, participating in hydrophobic core formation (Figures 2c &3a). As a result, the triangle-like conformation made by α3, α4, and the loop between them is stabilized. However, in ImB2, the connecting loop between α3 and α4 is not implicated in hydrophobic core formation. Instead the loop is located outside the four helix bundle structure. Consequently, α3 and α4 of ImB2 is arranged in a nearly parallel manner, extensively contacting with each other along the helical axis (Figure 3a). In conclusion, in spite of the overall similarity in structure, the difference of the C-terminal conformation and the local structure constructed by α3, α4, and the connecting loop between them can be used for the classification of pediocin-like immunity proteins.
Extreme C-terminal end of PedB is essential for immunity
The fact that PedB and EntA-im have almost identical four-helix bundle structures but their cognate bacteriocins are not identical (Figure 1a), led us to hypothesize that the flexible C-terminus region of PedB is critical for specific recognition of pediocin PP-1. Actually, according to the sequence alignment between PedB and EntA-im (Figure 1b), the sequence conservation of this region is much lower than that of other region. In order to verify the role of the C-terminal end, we introduced several shortened PedBs (PedB1–93, PedB1–96, PedB1–99, PedB1–102, PedB1–105, and PedB1–108) into L. sakei and tested the susceptibility to pediocin PP-1.
As shown in Figure 4a, deletion of last four residues (PedB1–108) resulted in severe loss of immunity compared to the wild type PedB, and deletion of seven residues (PedB1–105) almost completely abolished the activity. Since the flexible C-terminal end is not involved in the folding of PedB, the deletion cannot disturb the core structure of PedB. Actually, in the far-UV spectral region (190~250 nm), the CD spectra of PedB variants were highly similar to that of the wild type PedB (Figure 4b), which exhibits two negative bands at 208 and 222 nm supporting the all alpha helical structure of PedB [33]. Furthermore, in the near-UV spectral region (240~360 nm), PedB variants showed almost identical CD spectra to that of the wild type PedB (Figure 4c), indicating that PedB and PedB variants have very similar tertiary structure. Therefore, the mutant study indicates that the extreme C-terminus is crucial for the PedB immunity. Interestingly, in the flexible C-terminal end, several polar or charged residues are located from the position of 104 (Glu104, Thr105, Thr106, Asn107, Thr108, Ser111, and Gln112). The significant loss of immunity on the deletion of extreme C-terminus suggests that polar interactions mediated by these polar or charged residues might be involved in the specific recognition of pediocin PP-1.
Conclusion
We have determined the 1.6 Å crystal structure of PedB. The three pediocin-like immunity proteins whose structures are known (subgroup A and C) share a four helix bundle structure. In addition, the subgroup B immunity proteins, such as sakacin P and piscicolin 126 [23], are also predicted to consist of four helices by secondary structure prediction using the Jpred server [34] (data not shown), strongly suggesting that the four helix bundle is a conserved scaffold in the pediocin-like immunity proteins. The structural comparison among pediocin-like immunity proteins reveals that the C-terminal conformation and the local structure constructed by α3, α4, and the connecting loop between them exhibits a considerable disparity according to subgroup type, indicating that the pediocin-like immunity proteins could be classified into subgroups based on the structure as well as the sequence homology. The mode of action of pediocin-like immunity proteins still remains elusive. However, our results demonstrate that the extreme C-terminus of PedB with several charged residues is essential for immunity. Further analysis about what is the membrane-bound receptor for type IIa bacteriocins and how the recognition between the immunity proteins and cognate bacteriocins is achieved in a highly specific manner, is a great challenge in the future.
Methods
Bacterial strains and growth conditions
Pediocin PP-1 was produced by P. pentosaceus CBT8. L. sakei NCFB 2714 containing a plasmid with or without inserted PedB variants was used as an indicator strain for assaying immunity activity against pediocin PP-1. For overexpression and selenomethionine-labelling, E. coli strain BL21 (DE3) and B834 (DE3) (Novagen) were used respectively. P. pentosaceus and L. sakei were grown in MRS broth (Difco) at 30°C, and E. coli strains were grown in Luria-Bertani medium at 37°C. Chloramphenicol (Sigma) was used at 5 μg ml-1 for L. sakei and 34 μg ml-1 for E. coli. Ampicillin (Sigma) was used at 100 μg ml-1.
Construction of plasmids expressing PedB variants and transformation
In order to construct a plasmid expressing the wild type pedB gene under the control of the promoter of sodA, DNA fragments containing the pedB ORF were amplified by polymerase chain reaction (PCR) using two primers possessing Nde I and Bam HI site, respectively, and cloned into pGEMTeasy (Promega) generating pGEMT-pedB. Eco RI-Bam HI fragment of pGEMT-pedB was ligated into pGEX4T-1, generating pGEX-pedB. The sodA promoter was amplified by PCR using B. subtilis genomic DNA as a template and cloned into pGEMTeasy, generating pGEMT-psodA. This plasmid was digested with Sal I-Nde I and inserted into the pGEX-pedB, generating pGEX-psodA:pedB. Sal I-Bam HI fragment of this plasmid was ligated into pSET5s [35], generating pSET-psodA:pedB. As a control, Sal I-Eco RI fragment of pGEMT-psodA was also cloned to pSET5s, generating pSET-psodA. To construct plasmids for shortened PedB variants, each PedB variant was amplified by PCR with proper primer pairs containing stop codon at positions of 94, 97, 100, 103, 106, and 109, respectively. And then, 0.35 kb Nde I-Bam HI fragments of the PCR products were exchanged with pedB in pGEX-psodA:pedB. Finally, Sal I-Bam HI fragment of these plasmids were moved to pSET5s.
The expression constructs were electroporated into the L. sakei using Bio-Rad gene pulser as described by Aukrust et al. [36]. Briefly, L. sakei was made electrocompetent as follows. Cells were grown up to OD600 of approximately 0.5 in MRS broth with 2% (w/v) glycine, and then were washed twice with 1 mM MgCl2 followed by washing twice with 30% (w/v) polyethylene glycol 1500.
Protein purification, crystallization, and structure determination
PedB was expressed in E. coli and purified as described previously [37]. To ensure an anomalous signal in the X-ray diffraction experiment one leucine (Leu24) was mutated to methionine (L24M). For the selenomethionine labeling of PedB-L24M mutant, methionine auxotroph E. coli B834 (DE3) (Novagen) strain was used as a host for plasmid transformation. The selenomethionyl PedB-L24M protein was purified by an identical procedure to that for the wild-type PedB. The purified SeMet PedB-L24M was concentrated to approximately 10 mg ml-1 for crystallization.
Native PedB crystals (P 62; a = b = 62.2 and c = 39.9 Å, a monomer in asymmetric unit) were grown and a 1.6 Å data set was obtained with a Bruker Proteum 300 CCD at Beamline 6B at Pohang Light Source as described previously [37].
The selenomethionyl PedB-L24M crystals were produced under the condition containing 2.4 M ammonium sulfate, 0.3 M sodium chloride, and 0.1 M MES (pH 6.0). These crystals belonged to space group P 62 with cell parameters a = b = 62.6, and c = 39.8 Å, and contained one monomersper asymmetric unit. Diffraction data were processed and scaled with programs DENZO and SCALEPACK from the HKL program suite [38]. The structure was solved by the multiwavelength anomalous dispersion (MAD) method using three-wavelength data sets of the selenomethionyl L24M mutant crystal: the peak wavelength was at 0.97900 Å, and the inflection wavelength was at 0.97911 Å, and the remote wavelength was at 0.97137 Å. Two selenium sites were located, and phase refinement was done by using the programs SOLVE and RESOLVE [39, 40]. Initial phasing and model building was done with L24M MAD data set. Model building was done using the program QUANTA software (Molecular Simulations) and refined with CNS [41] to an Rfree of 0.276 and Rwork of 0.256 in the resolution range of 20-1.9 Å. The native PedB structure was solved by molecular replacement with CNS using the L24M structure as a search model, and refined to Rfree of 0.225 and Rwork of 0.192 in the resolution range of 20-1.6 Å. The refinement statistics for the native PedB structure are summarized in Table 1. The ideality of the model stereochemistry was verified by PROCHECK [42]. The Ramachandran plots indicate that 96.3% of non-glycine residues are in the most favored regions, and 2.4% in the additionally allowed regions, and 1.2% in the generously allowed regions. Coordinate has been deposited with the Protein Data Bank under the accession code 2IP6.
Determination of oligomeric state of PedB
Purified recombinant PedB and low molecular mass standards (Amersham Biosciences) were applied on a Superdex 75 16/60 column (Amersham Biosciences) preequilibrated with 20 mM Tris-HCl (pH 8.0) containing 150 mM NaCl. A standard curve was generated by plotting the logarithm of molecular mass of standard proteins against their Kav, where Kav = (Ve - Vo)/(Vt - Vo): Ve, elution volume; Vo, void volume; Vt, total bed volume. Kav of PedB determined by using the same column was compared to the profile of protein standards: bovine serum albumin (67 kDa), ovalbumin (43 kDa), chymotrypsinogen A (25 kDa), and ribonuclease A (13.7 kDa).
Purification of C-terminal shortened PedB mutants in E. coli
To construct plasmids for expression of shortened PedB variants in E. coli, each PedB variant was amplified by PCR with proper primer pairs containing stop codon at positions of 94, 97, 100, 103, 106, and 109, respectively. And then, 0.35 kb Bam HI-Sal I fragments of the PCR products were cloned into pGEX-4T-1. Each PedB variant was expressed and purified by an identical procedure to that for the wild type PedB.
CD Spectroscopy
All of the CD experiments were performed on a Jasco J-715 spectropolarimeter using a 0.2 cm path-length cell, with a 1 nm bandwidth and 4 s response time. Far- and near-UV CD spectra were collected from 250 to 190 nm and from 360 to 240 nm, respectively, with a scan speed of 100 nm/min and 1 nm step resolution. Three individual scans were added and averaged. A buffer containing 20 mM Tris-HCl (pH 7.4) and 150 mM NaCl was used for CD spectra.
Bacteriocin assay
For the bacteriocin assay, we used an ammonium sulphate-precipitated fermentate of P. pentosaceus containing pediocin PP-1. After the ammonium sulphate was added to the culture supernatant of P. pentosaceus to 80% saturation, the precipitate was dissolved in 10 mM sodium acetate pH 5.0. When this ammonium sulphate-precipitated fermentate was applied to the spot test against lawns of Listeria monocytogenes and Listeria innocua on agar media, it showed strong inhibitory effect (see Additional file 1). However, when proteinase K and chymotrypsin were treated to the fermentate, the clear zone disappeared (data not shown), indicating that the growth inhibition was due to pediocin PP-1 peptide.
Bacteriocin activity was measured using a microtiter plate assay system as described by Nissen-Meyer et al. [43]. Briefly, each well of a plate contained 100 μl of MRS medium mixed with ammonium sulphate-precipitated fermentate of P. pentosaceus at 1.5 fold dilution and L. sakei strain that is 1000 fold diluted at an OD600 of 1. This plate was incubated overnight at 30°C, and then the growth was measured at 595 nm using a microplate reader. The minimal inhibitory concentration (MIC) was defined as the concentration of bacteriocin that inhibited growth of the indicator strain by 50%. The MICs presented here are the results of three to four independent measurements. Transformed indicator strains were grown in the presence of 5 μg ml-1 chloramphenicol to maintain the plasmids in the cells. The immunity activity of PedB was calculated by comparing MIC values for a strain expressing the PedB variants with those for the strain possessing only the sodA promoter.
References
Guder A, Wiedemann I, Sahl HG: Posttranslationally modified bacteriocins-the lantibiotics. Biopolymers 2000, 55(1):62–73. 10.1002/1097-0282(2000)55:1<62::AID-BIP60>3.0.CO;2-Y
Papagianni M: Ribosomally synthesized peptides with antimicrobial properties: biosynthesis, structure, function, and applications. Biotechnol Adv 2003, 21(6):465–499. 10.1016/S0734-9750(03)00077-6
van Belkum MJ, Stiles ME: Nonlantibiotic antibacterial peptides from lactic acid bacteria. Nat Prod Rep 2000, 17(4):323–335. 10.1039/a801347k
Sahl HG, Jack RW, Bierbaum G: Biosynthesis and biological activities of lantibiotics with unique post-translational modifications. Eur J Biochem 1995, 230(3):827–853. 10.1111/j.1432-1033.1995.tb20627.x
Ennahar S, Sashihara T, Sonomoto K, Ishizaki A: Class IIa bacteriocins: biosynthesis, structure and activity. FEMS Microbiol Rev 2000, 24(1):85–106. 10.1111/j.1574-6976.2000.tb00534.x
Nissen-Meyer J, Nes IF: Ribosomally synthesized antimicrobial peptides: their function, structure, biogenesis, and mechanism of action. Arch Microbiol 1997, 167(2–3):67–77. 10.1007/s002030050418
Fimland G, Eijsink VG, Nissen-Meyer J: Mutational analysis of the role of tryptophan residues in an antimicrobial peptide. Biochemistry 2002, 41(30):9508–9515. 10.1021/bi025856q
Chikindas ML, Garcia-Garcera MJ, Driessen AJ, Ledeboer AM, Nissen-Meyer J, Nes IF, Abee T, Konings WN, Venema G: Pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0, forms hydrophilic pores in the cytoplasmic membrane of target cells. Appl Environ Microbiol 1993, 59(11):3577–3584.
Moll GN, Konings WN, Driessen AJ: Bacteriocins: mechanism of membrane insertion and pore formation. Antonie Van Leeuwenhoek 1999, 76(1–4):185–198. 10.1023/A:1002002718501
Uteng M, Hauge HH, Markwick PR, Fimland G, Mantzilas D, Nissen-Meyer J, Muhle-Goll C: Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide sakacin P and a sakacin P variant that is structurally stabilized by an inserted C-terminal disulfide bridge. Biochemistry 2003, 42(39):11417–11426. 10.1021/bi034572i
Fregeau Gallagher NL, Sailer M, Niemczura WP, Nakashima TT, Stiles ME, Vederas JC: Three-dimensional structure of leucocin A in trifluoroethanol and dodecylphosphocholine micelles: spatial location of residues critical for biological activity in type IIa bacteriocins from lactic acid bacteria. Biochemistry 1997, 36(49):15062–15072. 10.1021/bi971263h
Wang Y, Henz ME, Gallagher NL, Chai S, Gibbs AC, Yan LZ, Stiles ME, Wishart DS, Vederas JC: Solution structure of carnobacteriocin B2 and implications for structure-activity relationships among type IIa bacteriocins from lactic acid bacteria. Biochemistry 1999, 38(47):15438–15447. 10.1021/bi991351x
Chen Y, Ludescher RD, Montville TJ: Electrostatic interactions, but not the YGNGV consensus motif, govern the binding of pediocin PA-1 and its fragments to phospholipid vesicles. Appl Environ Microbiol 1997, 63(12):4770–4777.
Kazazic M, Nissen-Meyer J, Fimland G: Mutational analysis of the role of charged residues in target-cell binding, potency and specificity of the pediocin-like bacteriocin sakacin P. Microbiology 2002, 148(Pt 7):2019–2027.
Fimland G, Blingsmo OR, Sletten K, Jung G, Nes IF, Nissen-Meyer J: New biologically active hybrid bacteriocins constructed by combining regions from various pediocin-like bacteriocins: the C-terminal region is important for determining specificity. Appl Environ Microbiol 1996, 62(9):3313–3318.
Miller KW, Schamber R, Osmanagaoglu O, Ray B: Isolation and characterization of pediocin AcH chimeric protein mutants with altered bactericidal activity. Appl Environ Microbiol 1998, 64(6):1997–2005.
Axelsson L, Holck A: The genes involved in production of and immunity to sakacin A, a bacteriocin from Lactobacillus sake Lb706. J Bacteriol 1995, 177(8):2125–2137.
Dayem MA, Fleury Y, Devilliers G, Chaboisseau E, Girard R, Nicolas P, Delfour A: The putative immunity protein of the Gram-positive bacteria Leuconostoc mesenteroides is preferentially located in the cytoplasm compartment. FEMS Microbiol Lett 1996, 138(2–3):251–259. 10.1111/j.1574-6968.1996.tb08166.x
Huhne K, Axelsson L, Holck A, Krockel L: Analysis of the sakacin P gene cluster from Lactobacillus sake Lb674 and its expression in sakacin-negative Lb. sake strains. Microbiology 1996, 142: 1437–1448.
Quadri LE, Sailer M, Terebiznik MR, Roy KL, Vederas JC, Stiles ME: Characterization of the protein conferring immunity to the antimicrobial peptide carnobacteriocin B2 and expression of carnobacteriocins B2 and BM1. J Bacteriol 1995, 177(5):1144–1151.
Quadri LE, Kleerebezem M, Kuipers OP, de Vos WM, Roy KL, Vederas JC, Stiles ME: Characterization of a locus from Carnobacterium piscicola LV17B involved in bacteriocin production and immunity: evidence for global inducer-mediated transcriptional regulation. J Bacteriol 1997, 179(19):6163–6171.
Eijsink VG, Skeie M, Middelhoven PH, Brurberg MB, Nes IF: Comparative studies of class IIa bacteriocins of lactic acid bacteria. Appl Environ Microbiol 1998, 64(9):3275–3281.
Fimland G, Eijsink VG, Nissen-Meyer J: Comparative studies of immunity proteins of pediocin-like bacteriocins. Microbiology 2002, 148(Pt 11):3661–3670.
Johnsen L, Fimland G, Nissen-Meyer J: The C-terminal domain of pediocin-like antimicrobial peptides (class IIa bacteriocins) is involved in specific recognition of the C-terminal part of cognate immunity proteins and in determining the antimicrobial spectrum. J Biol Chem 2005, 280(10):9243–9250. 10.1074/jbc.M412712200
Johnsen L, Fimland G, Mantzilas D, Nissen-Meyer J: Structure-function analysis of immunity proteins of pediocin-like bacteriocins: C-terminal parts of immunity proteins are involved in specific recognition of cognate bacteriocins. Appl Environ Microbiol 2004, 70(5):2647–2652. 10.1128/AEM.70.5.2647-2652.2004
Venema K, Kok J, Marugg JD, Toonen MY, Ledeboer AM, Venema G, Chikindas ML: Functional analysis of the pediocin operon of Pediococcus acidilactici PAC1.0: PedB is the immunity protein and PedD is the precursor processing enzyme. Mol Microbiol 1995, 17(3):515–522. 10.1111/j.1365-2958.1995.mmi_17030515.x
Johnsen L, Dalhus B, Leiros I, Nissen-Meyer J: 1.6 Å crystal structure of EntA-im. A bacterial immunity protein conferring immunity to the antimicrobial activity of the pediocin-like bacteriocin enterocin A. J Biol Chem 2005, 280(19):19045–19050. 10.1074/jbc.M501386200
Sprules T, Kawulka KE, Vederas JC: NMR solution structure of ImB2, a protein conferring immunity to antimicrobial activity of the type IIa bacteriocin, carnobacteriocin B2. Biochemistry 2004, 43(37):11740–11749. 10.1021/bi048854+
Marugg JD, Gonzalez CF, Kunka BS, Ledeboer AM, Pucci MJ, Toonen MY, Walker SA, Zoetmulder LC, Vandenbergh PA: Cloning, expression, and nucleotide sequence of genes involved in production of pediocin PA-1, and bacteriocin from Pediococcus acidilactici PAC1.0. Appl Environ Microbiol 1992, 58(8):2360–2367.
Inaoka T, Matsumura Y, Tsuchido T: Molecular cloning and nucleotide sequence of the superoxide dismutase gene and characterization of its product from Bacillus subtilis . J Bacteriol 1998, 180(14):3697–3703.
Inaoka T, Matsumura Y, Tsuchido T: SodA and manganese are essential for resistance to oxidative stress in growing and sporulating cells of Bacillus subtilis . J Bacteriol 1999, 181(6):1939–1943.
Chothia C, Lesk AM: The relation between the divergence of sequence and structure in proteins. EMBO J 1986, 5(4):823–826.
Greenfield N, Fasman GD: Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry 1969, 8: 4108–4116. 10.1021/bi00838a031
Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ: JPred: a consensus secondary structure prediction server. Bioinformatics 1998, 14(10):892–893. 10.1093/bioinformatics/14.10.892
Takamatsu D, Osaki M, Sekizaki T: Thermosensitive suicide vectors for gene replacement in Streptococcus suis . Plasmid 2001, 46(2):140–148. 10.1006/plas.2001.1532
Aukrust TW, Brurberg MB, Nes IF: Transformation of Lactobacillus by electroporation. Methods Mol Biol 1995, 47: 201–208.
Kim IK, Kim MK, Yim HS, Cha SS, Kang SO: Crystallization and preliminary X-ray crystallographic analysis of the pediocin immunity protein (PedB) from Pediococcus pentosaceus at 1.35 Å resolution. Biochim Biophys Acta 2005, 1751(2):205–208.
Otwinowski Z, Minor W: Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 1997, 276: 307–326.
Terwilliger TC: Reciprocal-space solvent flattening. Acta Crystallogr Sect D Biol Crystallogr 1999, 55(11):1863–1871. 10.1107/S0907444999010033
Terwilliger TC, Berendzen J: Discrimination of solvent from protein regions in native Fouriers as a means of evaluating heavy-atom solutions in the MIR and MAD methods. Acta Crystallogr Sect D Biol Crystallogr 1999, 55(Pt 2):501–505. 10.1107/S0907444998012657
Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al.: Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr Sect D Biol Crystallogr 1998, 54(Pt 5):905–921. 10.1107/S0907444998003254
Laskowski RA, MacArthur MW, Moss DS, Thornton JM: PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogra 1993, 26: 283–291. 10.1107/S0021889892009944
Nissen-Meyer J, Holo H, Havarstein LS, Sletten K, Nes IF: A novel lactococcal bacteriocin whose activity depends on the complementary action of two peptides. J Bacteriol 1992, 174(17):5686–5692.
Acknowledgements
Work performed at Seoul National University was supported by the 21C Frontier Microbial Genomics and Application Center Program, Ministry of Science & Technology(Grant MG02-0201-001-1-0-0), Republic of Korea and by research fellowship of the BK21 project and work performed at Pohang Accelerator Laboratory was supported by a grant from the Functional Proteomics Center, the 21C Frontier Research & Development Program of the Korea Ministry of Science and Technology, Republic of Korea and in part by a research grant from the National Institute of Health (NIH) of KCDC in Republic of Korea.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Authors' contributions
IK carried out the protein expression, purification, and crystallization experiments, participated in structure determination, construction of PedB-expressing plasmids, and bacteriocin assay, and wrote the initial version of the manuscript. MK carried out the site-directed mutagenesis, mutant purification, crystallization and participated in data analysis, structure determination, and electroporation. JK participated in the electroporation and bacteriocin assay. HY participated in data analysis and design of this study. SC and SK conceived the project, supervised the project, participated in its design and coordination, and helped to draft the manuscript. All authors read and approved the final manuscript.
In-Kwon Kim, Min-Kyu Kim contributed equally to this work.
Electronic supplementary material
12900_2006_116_MOESM1_ESM.pdf
Additional file 1: Detection of pediocin PP-1 activity in the absence and presence of PedB. The ammonium sulphate-precipitated fermentate was applied to the spot test against lawns of L. innocua and L. monocytogenes on agar media. (PDF 36 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Kim, IK., Kim, MK., Kim, JH. et al. High resolution crystal structure of PedB: a structural basis for the classification of pediocin-like immunity proteins. BMC Struct Biol 7, 35 (2007). https://doi.org/10.1186/1472-6807-7-35
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1472-6807-7-35