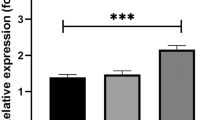
Abstract
Background
Knee osteoarthritis (OA) is a complex disease involving both biomechanical and metabolic factors that alter the tissue homeostasis of articular cartilage and subchondral bone. The catabolic activities of extracellular matrix degradation products, especially fibronectin (FN), have been implicated in mediating cartilage degradation. Chondrocytes express several members of the integrin family which can serve as receptors for FN including integrins α5β1, αvβ3, and αvβ5. The purpose of this study was to determine whether polymorphisms in the FN (FN-1) and integrin genes are markers of susceptibility to, or severity of, knee OA in a Han Chinese population.
Methods
Two independent case–control studies were conducted on 928 patients with knee OA and 693 healthy controls. Ten single nucleotide polymorphisms (SNPs) of FN-1 and the integrin αV gene (ITGAV) were detected using the ABI 7500 real-time PCR system.
Results
The AT heterozygote in FN-1 (rs940739A/T) was found to be significantly associated with knee OA (adjusted OR = 1.44; 95% CI = 1.16–1.80) in both stages of the study. FN-1 rs6725958C/A and ITGAV rs10174098A/G SNPs were only associated with knee OA when both study groups were combined. Stratifying the participants by Kellgren-Lawrence (KL) score identified significant differences in the FN-1 rs6725958C/A and rs940739 A/T genotypes between patients with grade 4 OA and controls. Haplotype analyses revealed that TGA and TAA were associated with a higher risk of OA, and that TAG conferred a lower risk of knee OA in the combined population.
Conclusions
Our study suggests that the FN-1 rs940739A/T polymorphism may be an important risk factor of genetic susceptibility to knee OA in the Han Chinese population.
Similar content being viewed by others
Background
Osteoarthritis (OA) is a degenerative joint disease that progressively causes loss of joint function and is a leading cause of disability and impaired quality of life among the elderly in developed countries. It is characterized by degeneration and the progressive loss of articular cartilage with pathological changes in bone, synovium, and other soft tissues of affected joints [1]. A number of factors, such as genetic predisposition, aging, obesity, inflammation, and excessive mechanical loading, are recognized to contribute to OA onset and progression [2]. As such, OA is a complex disease involving both biomechanical and metabolic factors that alter the tissue homeostasis of articular cartilage and subchondral bone [3].
The catabolic activities of matrix degradation products, including fibronectin fragments (FN-fs), have been implicated in mediating cartilage degradation [4, 5]. At least some of these fragments appear to act via an integrin-dependent mechanism [6]. Chondrocytes express several members of the integrin family that can serve as FN receptors, including α5β1, αvβ3, and αvβ5 [7, 8].
FN is encoded by FN-1, which is located on chromosome 2q34-36, is over 75 kb in length and is composed of 50 exons. FN is a multifunctional glycoprotein that is involved in a wide range of biological processes such as cell migration, wound healing, angiogenesis, and differentiation. FN exists in two major forms. The soluble form is found in the plasma, while the insoluble form is present in tissues, including at low levels in the extracellular matrix (ECM) of normal cartilage [9]. Alternative splicing of two type III exons, known as Extra Domains A and B, and a variable region result in the production of up to 20 different FN variants [10]. Growing evidence suggests that these isoforms affect the function of FN and FN-fs, including cartilage breakdown in OA [11].
Integrins are cell surface receptors composed of α and β chains. The integrin family consists of at least 24 different heterodimers that are involved in cell–cell and cell–ECM adhesion [12]. Integrin αV (ITGAV) is one of the more promiscuous alpha integrins associating with five different beta subunits including β1, β3, β5, β6, and β8 [13]. Expression of ITGAV appears to be upregulated in osteoarthritic chondrocytes [14].
Single nucleotide polymorphisms (SNPs) represent variations in the genome between individuals. Several studies have demonstrated that a number of these polymorphisms are associated with OA and may contribute to the genetic risk of developing the disease [15–17]. Because FN-1 and ITGAV polymorphisms may influence gene expression or modify the biological activities of the functional protein, we set out to test the hypothesis that such polymorphisms influence susceptibility and the severity of knee OA.
Methods
A total of 928 patients with knee OA and 693 healthy individuals were recruited for two independent studies carried out between July 2008 and December 2009 (the first study conducted in Miaoli, which is located in a largely rural area of China) and between March 2010 and June 2011 (the second study conducted in the centrally located Chinese city of Taipei). The first study recruited and genotyped 403 knee OA patients (65.4% female) and 314 controls (53.8% female) to identify knee OA-associated SNPs and haplotypes. The second study recruited and genotyped 525 knee OA patients (65.6% female) and 379 healthy controls (58.6% female).
Disease severity in the knee OA populations, who were attending an orthopedic hospital, was assessed by Kellgren-Lawrence (KL) grading. All patients had a KL score ≥ 2. Other etiologies of knee joint disease such as inflammatory arthritis, post-traumatic or post-septic arthritis, and skeletal or developmental dysplasia were excluded from the study. Healthy control subjects had no signs or symptoms of joint disease (pain, swelling, tenderness, or restriction of movement) and standard x-rays of the knee joints confirmed an absence of OA. This study was reviewed and approved by the institutional ethical committee of Taipei Medical University Hospital and Tri-Service General Hospital (CRC-01-10-03 and TSGH-100-05-023). All clinical and biological samples were collected and DNA was genotyped following approval by this committee. Written informed consent was obtained from all participants after the study had been fully explained to them.
Selection and genotyping of polymorphisms
We selected FN-1 and ITGAV as candidate genes based on the published literature [18, 19]. SNP genotype information was downloaded from the HapMap database (http://hapmap.ncbi.nlm.nih.gov/) and The National Center for Biotechnology Information dbSNP database (http://www.ncbi.nlm.nih.gov/snp) to select the most representative SNPs by capturing the majority of genetic variation. Using the tagger program implemented in Haploview 4.0, tag SNPs across FN-1 and ITGAV were selected on the basis of linkage disequilibrium patterns observed in the Han Chinese samples genotyped as part of the International HapMap Project. Only SNPs with a minor allele frequency greater than 5% in HapMap were considered. Six SNPs (rs10202709, rs6725958, rs940739, rs2304573, rs11651, and rs3796123) in FN-1 and four SNPs (rs3911238, rs10174098, rs3738929, and rs1448427) in ITGAV were included to capture as much variation as possible. These 10 tag SNPs captured all alleles with an r2 of at least 0.8.
Genomic DNA was extracted from the peripheral blood of patients and controls using the QIAamp DNA Blood Mini Kit (QIAGEN Inc., Hilden, Germany) according to the manufacturer’s instructions. Genotyping was performed using the TaqMan® SNP Genotyping Assay (Applied Biosystems, Foster City, CA, USA) with the following cycling conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min, with a 1 min extension at 25°C following the last cycle. Genotyping was performed by laboratory personnel blinded to case status and 10% of the samples were randomly selected for repeated testing to validate genotyping procedures. Two authors independently reviewed the genotyping results, data entry, and statistical analyses.
Statistical analysis
Statistical analysis was performed using SPSS for Windows version 18.0 software (SPSS, Chicago, IL, USA), and results were considered statistically significant where the two-tailed p-value was less than 0.05. SNP deviations from the Hardy-Weinberg equilibrium (HWE) in control samples were assessed using the standard chi-squared (χ2) test. The allele and genotype frequencies of OA patients and control subjects were compared using χ2 statistics or Fisher’s exact test as appropriate. The demographics were evaluated by the Student’s t-test or Mann–Whitney U test for continuous variables and expressed as means ± standard deviation (SD). Logistic regression was used to estimate crude and adjusted age, gender, and body mass index (BMI) odd ratios (ORs) and 95% confidence intervals (CIs) as a measure of association with the risk of OA.
Linkage disequilibrium and haplotype analyses were performed using Haploview software (http://www.broad.mit.edu/mpg/haploview/) [20]. Because the frequency distributions of demographic characteristics and genotypic and allelic distributions of the 10 SNPs were similar between the two study cohorts, all data were pooled to analyze the association between SNPs and knee OA. In the absence of interstudy heterogeneity within samples, we also constructed a Mantel-Haenszel meta-analysis of sample data to assess the overall evidence of association. The Mantel-Haenszel χ2 test and estimate of the OR were computed with or without the inclusion of covariates using R, version 3.0.2, with the “metafor” and “meta” packages.
The assumption of heterogeneity for each analysis was tested using the DerSimonian-Laird method. The level of significance was determined by Bonferroni’s method for correcting multiple testing errors. For the 10 selected SNPs, a p-value < 0.005 (0.05 divided by 10) was considered statistically significant. In this study, power estimation was performed using CaTS (http://www.sph.umich.edu/csg/abecasis/CaTS/) and is summarized in Additional file 1: Table S1.
Results
Characteristics of study subjects
The frequency distributions of demographic characteristics between the 928 knee OA cases and 693 healthy controls in the two independent studies are shown in Table 1. The average age of knee OA cases was significantly higher than the control population. The proportion of females in the knee OA cases (65.5%) was significantly higher than in the healthy controls (56.4%). BMI was also slightly higher in the knee OA cases.
Association analyses of FN-1 polymorphisms with susceptibility to OA
Table 2 shows the genotype distributions and allele frequencies of FN-1 polymorphisms in knee OA and control subjects from both studies. The genotype frequencies of all six FN-1 SNPs were in HWE (p > 0.05). In study 1, the genotype distribution of FN-1 rs940739A/T was significantly different between knee OA patients and healthy controls (p < 0.05). When the FN-1 rs940739AA genotype was used as the reference group, the rs940739AT genotype appeared to be associated with a higher risk for knee OA (adjusted OR = 1.44, 95% CI = 1.03–2.01). To determine whether the results could be replicated, we genotyped the six SNPs in the second study population. In this study group, rs940739AT also differed significantly in genotype distribution between the knee OA cases and healthy controls (adjusted OR = 1.48, 95% CI = 1.10–1.99).
The other FN-1 SNPs (rs10202709G/A, rs6725958C/A, rs2304573G/A, rs11651A/G, and rs3796123T/A) demonstrated no significant genotypic or allelic association between OA cases and healthy controls in either of the two study groups (Table 2).
Association analyses for FN-1 polymorphisms in the combined studies
When data from the two independent studies were combined, the association of the rs940739A/T polymorphism with knee OA was maintained (adjusted OR = 1.44, 95% CI = 1.16–1.80, p = 0.001). Additionally, the minor allele of FN-1 rs6725958C/A appeared to be associated with a significantly higher risk of knee OA (adjusted OR = 1.17, 95% CI = 1.01–1.80, p = 0.033) (Table 3). After correcting for multiple comparisons, the FN-1 rs940739TA genotype had a higher risk for OA, but the minor allele of FN-1 rs6725958C/A was not significant.
Association analyses for ITGAV polymorphisms in the combined studies
Genotypic and allelic distributions of the four ITGAV SNPs and their associations with knee OA risk are shown in Table 4. No deviation from the HWE was observed in the controls (p > 0.05). There was no significant genotypic or allelic association identified between the knee OA cases and healthy controls with the four ITGAV SNPs in both study cohorts. However, when data from the two independent studies were combined, rs10174098A/G was associated with a lower risk of knee OA (adjusted OR = 0.77, 95% CI = 0.62–0.96, p = 0.020).
In males, significant associations were observed between ITGAV rs3738919C/A and OA (Additional file 1: Table S4), but no associations were found between OA and FN-1 polymorphisms among males (Additional file 1: Table S2). In females, genotypic and allelic frequencies of FN-1 rs6725958C/A and rs940739A/T and ITGAV rs10174098A/G polymorphisms differed significantly between knee OA cases and healthy controls (Additional file 1: Tables S3 and S4). Additionally, the dominant mode of FN-1 rs640739 also appeared to be associated with a significantly higher risk of knee OA (adjusted OR = 1.41, 95% CI = 1.14–1.74, p = 0.002) (Additional file 1: Table S5).
In the recessive mode, the 10 FN-1 and ITGAV SNPs showed no significance (Additional file 1: Table S6). After correcting for multiple comparisons, no SNPs appeared to be associated with OA. We further analyzed interactions between FN-1 rs940739A/T and obesity, but observed no significant interaction between the two (p for interaction = 0.15 for the interaction term, Additional file 1: Table S7).
Stratification analysis according to disease severity
Stratifying the patients by KL score identified significant differences in FN-1 rs6725958C/A alleles and genotypes between patients with both grade 3 and 4 knee OA and controls (AA/CC, adjusted OR = 2.11, 95% CI = 1.45–3.06; adjusted OR = 0.54, 95% CI = 0.36–0.82; CA/CC, adjusted OR = 1.58, 95% CI = 1.16–2.16). FN-1 rs940739A/T was also associated with grade 4 knee OA (AT/AA, adjusted OR = 2.07, 95% CI = 1.59–2.69) (Table 5).
Haplotype analysis of FN-1
Linkage disequilibrium analysis confirmed disequilibrium between SNPs rs2304573 and rs11651 (r2 = 0.73; data not shown). The haplotype analysis of FN-1 polymorphisms in knee OA cases and control subjects is shown in Table 6. The frequency of haplotype AAG was 6.6% in knee OA patients compared with 10.1% in controls (OR = 0.63, 95% CI = 0.49–0.81, p < 0.001). By contrast, haplotypes TGA and TAA were more common in knee OA patients (4.7% and 2.2%, respectively) than controls (2.4% and 1.0%, respectively) (OR = 2.03 and 2.12, 95% CI = 1.35–3.06 and 1.16–3.89, p < 0.001 and = 0.013, respectively).
Discussion
After conducting two independent studies involving the genotyping and evaluation of disease severity of 928 knee OA patients and 693 healthy controls from a Han Chinese population, we have identified an association between the FN-1 rs940739A/T SNP and OA. When data from both studies were combined, we also identified an association between the ITGAV rs10174098A/G SNP and knee OA.
Several recent studies have identified FN-1 mutations and polymorphisms that affect host susceptibility to a variety of diseases including calcium oxalate stone disease [21], schizophrenia [22], lung cancer [23], and fibrosing alveolitis in systemic sclerosis [24]. However, no previous studies have reported on an association between FN-1 polymorphisms and knee OA. The current study has, for the first time, revealed that the FN-1 intronic polymorphism rs940739A/T is associated with knee OA. SNPs in introns are known to influence the regulation of transcription, splicing, and other aspects of RNA processing or stability [25, 26]. For instance, the intronic SNPs in FTO [27], COL1A1 [28], and BANK1 [29] affect primary transcript levels while the risk allele of the intronic SNP in the prothrombin gene may influence splicing efficiency [30]. However, it is currently unclear whether the intronic SNP identified as being associated with knee OA in the current study performs a functional role by exerting a direct effect on FN-1 expression or whether it is in linkage disequilibrium with another functional SNP.
The role of FN and FN-fs in the development of knee OA is well recognized and supported by a number of studies. FN-fs, the central cell-, NH2-terminal heparin-, and NH2-terminal gelatin-binding fragments of FN have been shown to stimulate cartilage chondrolysis by inhibiting anabolic activity while increasing production of matrix metalloproteinases [31]. Importantly, FN-1 expression appears to be differentially regulated in monolayer cultured chondrocytes from OA and normal donors [32].
When we analyzed the association of SNP expression with the radiological grade of knee OA, a significant relationship was seen in patients with KL grade 4 knee OA with genotypes rs6725958C/A and rs940739A/T, indicating that these FN-1 polymorphisms are associated with advanced knee OA in the current Han Chinese population. The reasons for these associations are not clear. Genetic risk factors can influence the risk of development and progression of knee OA at various stages during the course of the disease. Because FN-1 polymorphisms could contribute to the risk of OA at different phases or throughout the disease process, further functional studies on such polymorphisms are required.
Haplotype analysis demonstrated an association between FN-1 and knee OA. Our data revealed that patients carrying TGA or TAA haplotypes had a higher risk of developing knee OA than those without, suggesting that multiple variants in FN-1 affect the risk. The FN-1 haplotype has previously been shown to have a significant influence on whether a fetus is at risk of being small-for-gestational-age [33], indicating that haplotype status could be important in tissue homeostasis.
Fibronectin and Fn-fs interact with chondrocytes and other joint tissue cells through integrins by which they can regulate cell function through a variety of intracellular signal cascades [5, 34, 35]. Although the α5β1 integrin is the major fibronectin receptor expressed by chondrocytes [7], αV is also abundantly expressed by articular chondrocytes; however, its biological significance in cartilage has been less well studied. ITGAV polymorphisms are associated with a number of conditions including rheumatoid arthritis [36], chronic hepatitis B virus infection [37], and primary biliary cirrhosis [38]. In the current study, the ITGAV SNP rs10174098A/G was only associated with knee OA when both study data were combined. Thus it is not entirely clear whether this weaker association with OA is genuine. Further independent studies need to be undertaken to clarify the situation.
Conclusion
The present study suggests that there is a significant association with the FN-1 polymorphism rs940739A/T and susceptibility to knee OA in a Chinese Han population. Such polymorphisms may explain why some individuals are at a higher risk for knee OA. Further studies on other larger populations combined with in vitro functional analyses are now crucial to elucidate the effects of the polymorphism on knee OA pathogenesis and susceptibility.
References
Litwic A, Edwards MH, Dennison EM, Cooper C: Epidemiology and burden of osteoarthritis. Br Med Bull. 2013, 105: 185-199. 10.1093/bmb/lds038.
Neogi T, Zhang Y: Epidemiology of osteoarthritis. Rheum Dis Clin North Am. 2013, 39: 1-19. 10.1016/j.rdc.2012.10.004.
Kuettner KE, Cole AA: Cartilage degeneration in different human joints. Osteoarthritis Cartilage. 2005, 13: 93-103. 10.1016/j.joca.2004.11.006.
Yasuda T: Cartilage destruction by matrix degradation products. Mod Rheumatol. 2006, 16: 197-205. 10.3109/s10165-006-0490-6.
Sofat N: Analysing the role of endogenous matrix molecules in the development of osteoarthritis. Int J Exp Pathol. 2009, 90: 463-479. 10.1111/j.1365-2613.2009.00676.x.
Werb Z, Tremble PM, Behrendtsen O, Crowley E, Damsky CH: Signal transduction through the fibronectin receptor induces collagenase and stromelysin gene expression. J Cell Biol. 1989, 109: 877-889. 10.1083/jcb.109.2.877.
Woods VL, Schreck PJ, Gesink DS, Pacheco HO, Amiel D, Akeson WH, Lotz M: Integrin expression by human articular chondrocytes. Arthritis Rheum. 1994, 37: 537-544. 10.1002/art.1780370414.
Salter DM, Hughes DE, Simpson R, Gardner DL: Integrin expression by human articular chondrocytes. Br J Rheumatol. 1992, 31: 231-234. 10.1093/rheumatology/31.4.231.
Rencic A, Gehris AL, Lewis SD, Hume EL, Bennett VD: Splicing patterns of fibronectin mRNA from normal and osteoarthritic human articular cartilage. Osteoarthritis Cartilage. 1995, 3: 187-196. 10.1016/S1063-4584(05)80053-6.
White ES, Baralle FE, Muro AF: New insights into form and function of fibronectin splice variants. J Pathol. 2008, 216: 1-14. 10.1002/path.2388.
Homandberg GA, Suppl 391: Cartilage damage by matrix degradation products: fibronectin fragments. Clin Orthop Relat Res. 2001, Oct, (Supp391): 100-107.
Calderwood DA: Integrin activation. J Cell Sci. 2004, 117: 657-666. 10.1242/jcs.01014.
Sims MA, Field SD, Barnes MR, Shaikh N, Ellington K, Murphy KE, Spurr N, Campbell DA: Cloning and characterisation of ITGAV, the genomic sequence for human cell adhesion protein (vitronectin) receptor alpha subunit, CD51. Cytogenet Cell Genet. 2000, 89: 268-271. 10.1159/000015631.
Iliopoulos D, Malizos KN, Oikonomou P, Tsezou A: Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS One. 2008, 3: e3740-10.1371/journal.pone.0003740.
Zeggini E, Panoutsopoulou K, Southam L, Rayner NW, Day-Williams AG, Lopes MC, Boraska V, Esko T, Evangelou E, Hoffman A, Houwing-Duistermaat JJ, Ingvarsson T, Jonsdottir I, Jonnson H, Kerkhof HJ, Kloppenburg M, Bos SD, Mangino M, Metrustry S, Slagboom PE, Thorleifsson G, Raine EV, Ratnayake M, Ricketts M, Beazley C, Blackburn H, Bumpstead S, Elliott KS, arcOGEN Consortium, arcOGEN Collaborators, et al: Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet. 2012, 380: 815-823.
Reynard LN, Loughlin J: Genetics and epigenetics of osteoarthritis. Maturitas. 2012, 71: 200-204. 10.1016/j.maturitas.2011.12.001.
Valdes AM, Spector TD: The genetic epidemiology of osteoarthritis. Curr Opin Rheumatol. 2010, 22: 139-143. 10.1097/BOR.0b013e3283367a6e.
Pulai JI, Del Carlo M, Loeser RF: The alpha5beta1 integrin provides matrix survival signals for normal and osteoarthritic human articular chondrocytes in vitro. Arthritis Rheum. 2002, 46: 1528-1535. 10.1002/art.10334.
Kim SJ, Kim EJ, Kim YH, Hahn SB, Lee JW: The modulation of integrin expression by the extracellular matrix in articular chondrocytes. Yonsei Med J. 2003, 44: 493-501.
Barrett JC, Fry B, Maller J, Daly MJ: Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005, 21: 263-265. 10.1093/bioinformatics/bth457.
Onaran M, Yilmaz A, Sen I, Ergun MA, Camtosun A, Kupeli B, Menevse S, Bozkirli I: A HindIII polymorphism of fibronectin gene is associated with nephrolithiasis. Urology. 2009, 74: 1004-1007. 10.1016/j.urology.2009.05.010.
Nakata K, Ujike H, Sakai A, Takaki M, Imamura T, Tanaka Y, Kuroda S: Association study between the fibronectin gene and schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2003, 116B: 41-44. 10.1002/ajmg.b.10796.
Siemianowicz K, Gminski J, Francuz T, Syzdol M, Polanska D, Machalski M, Brulinski K, Magiera-Molendowska H: Fibronectin gene polymorphism in patients with lung cancer. Oncol Rep. 2001, 8: 1289-1292.
Avila JJ, Lympany PA, Pantelidis P, Welsh KI, Black CM, Du Bois RM: Fibronectin gene polymorphisms associated with fibrosing alveolitis in systemic sclerosis. Am J Respir Cell Mol Biol. 1999, 20: 106-112. 10.1165/ajrcmb.20.1.3232.
Birney E, Lieb JD, Furey TS, Crawford GE, Iyer VR: Allele-specific and heritable chromatin signatures in humans. Hum Mol Genet. 2010, 19: R204-R209. 10.1093/hmg/ddq404.
Cheung VG, Spielman RS: Genetics of human gene expression: mapping DNA variants that influence gene expression. Nat Rev Genet. 2009, 10: 595-604. 10.1038/nrg2630.
Berulava T, Horsthemke B: The obesity-associated SNPs in intron 1 of the FTO gene affect primary transcript levels. Eur J Hum Genet. 2010, 18: 1054-1056. 10.1038/ejhg.2010.71.
Mann V, Hobson EE, Li B, Stewart TL, Grant SF, Robins SP, Aspden RM, Ralston SH: A COL1A1 Sp1 binding site polymorphism predisposes to osteoporotic fracture by affecting bone density and quality. J Clin Invest. 2001, 107: 899-907. 10.1172/JCI10347.
Kozyrev SV, Abelson AK, Wojcik J, Zaghlool A, Linga Reddy MV, Sanchez E, Gunnarsson I, Svenungsson E, Sturfelt G, Jönsen A, Truedsson L, Pons-Estel BA, Witte T, D'Alfonso S, Barizzone N, Danieli MG, Gutierrez C, Suarez A, Junker P, Laustrup H, González-Escribano MF, Martin J, Abderrahim H, Alarcón-Riquelme ME: Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat Genet. 2008, 40: 211-216. 10.1038/ng.79.
Von Ahsen N, Oellerich M: The intronic prothrombin 19911A > G polymorphism influences splicing efficiency and modulates effects of the 20210G > A polymorphism on mRNA amount and expression in a stable reporter gene assay system. Blood. 2004, 103: 586-593. 10.1182/blood-2003-02-0419.
Homandberg GA, Costa V, Wen C: Fibronectin fragments active in chondrocytic chondrolysis can be chemically cross-linked to the alpha5 integrin receptor subunit. Osteoarthritis Cartilage. 2002, 10: 938-949. 10.1053/joca.2002.0854.
Dehne T, Karlsson C, Ringe J, Sittinger M, Lindahl A: Chondrogenic differentiation potential of osteoarthritic chondrocytes and their possible use in matrix-associated autologous chondrocyte transplantation. Arthritis Res Ther. 2009, 11: R133-10.1186/ar2800.
Edwards DR, Romero R, Kusanovic JP, Hassan SS, Mazaki-Tovi S, Vaisbuch E, Kim CJ, Erez O, Chaiworapongsa T, Pearce BD, Bartlett J, Friel LA, Salisbury BA, Anant MK, Vovis GF, Lee MS, Gomez R, Behnke E, Oyarzun E, Tromp G, Menon R, Williams SM: Polymorphisms in maternal and fetal genes encoding for proteins involved in extracellular matrix metabolism alter the risk for small-for-gestational-age. J Matern Fetal Neonatal Med. 2011, 24: 362-380. 10.3109/14767058.2010.497572.
Kostopoulou F, Gkretsi V, Malizos KN, Iliopoulos D, Oikonomou P, Poultsides L, Tsezou A: Central role of SREBP-2 in the pathogenesis of osteoarthritis. PLoS One. 2012, 7: e35753-10.1371/journal.pone.0035753.
Guilak F: Biomechanical factors in osteoarthritis. Best Pract Res Clin Rheumatol. 2011, 25: 815-823. 10.1016/j.berh.2011.11.013.
Hollis-Moffatt JE, Rowley KA, Phipps-Green AJ, Merriman ME, Dalbeth N, Gow P, Harrison AA, Highton J, Jones PB, Stamp LK, Harrison P, Wordsworth BP, Merriman TR: The ITGAV rs3738919 variant and susceptibility to rheumatoid arthritis in four Caucasian sample sets. Arthritis Res Ther. 2009, 11: R152-10.1186/ar2828.
Lee SK, Kim MH, Cheong JY, Cho SW, Yang SJ, Kwack K: Integrin alpha V polymorphisms and haplotypes in a Korean population are associated with susceptibility to chronic hepatitis and hepatocellular carcinoma. Liver Int. 2009, 29: 187-195. 10.1111/j.1478-3231.2008.01843.x.
Inamine T, Nakamura M, Kawauchi A, Shirakawa Y, Hashiguchi H, Aiba Y, Taketomi A, Shirabe K, Nakamuta M, Hayashi S, Saoshiro T, Komori A, Yatsuhashi H, Kondo S, Omagari K, Maehara Y, Ishibashi H, Tsukamoto K, PBC Study Group in NHOSLJ: A polymorphism in the integrin alphaV subunit gene affects the progression of primary biliary cirrhosis in Japanese patients. J Gastroenterol. 2011, 46: 676-686. 10.1007/s00535-010-0351-0.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2474/15/173/prepub
Acknowledgements
This study was supported by grants from the National Science Council, National Defense Medical Center and Tri-Service General Hospital, Taiwan (S.L.S.: NSC99-2314-B-016-001; H.S.L.: TSGH-C102-068, MAB-102-57, NSC100-2320-B-016-006; C.H.L.: NSC101-2314-B -038-004, NSC100-2314-B-038-04).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
HYY conceived, designed, and performed the experiments, analyzed the data, and wrote the paper. SLS conceived and designed the experiments, analyzed the data, and contributed reagents/materials/analysis tools. HSL conceived, designed and performed the experiments, contributed reagents/materials/analysis tools, and wrote the paper. CHL conceived, designed and performed the experiments, contributed reagents/materials/analysis tools, and wrote the paper. YJP performed the experiments, and analyzed the data. CCW performed the experiments, and analyzed the data. DMS wrote the paper. All authors critically revised the manuscript and approved the final version.
Electronic supplementary material
12891_2013_2137_MOESM1_ESM.doc
Additional file 1: Table S1: Estimation of statistical power for the present study. Table S2. Genotype distributions and allele frequencies for the FN gene polymorphisms in male OA patients and healthy control groups. Table S3. Genotype distributions and allele frequencies for the FN gene polymorphisms in female OA patients and healthy control groups. Table S4. Genotype distributions and allele frequencies of the ITGAV gene by gender. Table S5. Analyses of the association of 10 SNPs in FN and ITGAV gene with OA (dominant model). Table S6. Analyses of the association of 10 SNPs in FN and ITGAV gene with OA (recessive model). Table S7. Joint effects of FN rs940739 and obesity among 928 cases of OA and 693 control subjects. (DOC 208 KB)
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Yang, HY., Su, SL., Peng, YJ. et al. An intron polymorphism of the fibronectin gene is associated with end-stage knee osteoarthritis in a Han Chinese population: two independent case- control studies. BMC Musculoskelet Disord 15, 173 (2014). https://doi.org/10.1186/1471-2474-15-173
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2474-15-173