Abstract
Background
Infections in newborns remain one of the most significant problems in modern medicine. Escherichia coli is an important cause of neonatal bloodstream and respiratory tract infections and is associated with high mortality. The aim of our study was to investigate the epidemiology of E. coli infection in Polish neonatal intensive care units (NICUs) and resistance to antibiotics, with particular reference to the safety of very low birth weight infants.
Methods
Continuous prospective infection surveillance was conducted in 2009–2012 in five NICUs, including 1,768 newborns whose birth weight was <1.5 kg. Escherichia coli isolates from different diagnostic specimens including blood, tracheal/bronchial secretions and others were collected. All isolates were tested using disk diffusion antimicrobial susceptibility methods. Pulsed-field gel electrophoresis was used to determine the possible horizontal transfer of E. coli among patients.
Results
The incidence of E. coli infections was 5.4% and 2.0/1,000 patient-days. The occurrence of E. coli infections depended significantly on the NICU and varied between 3.9% and 17.9%. Multivariate analysis that took into account the combined effect of demographic data (gender, gestational age and birth weight) and place of birth showed that only the place of hospitalisation had a significant effect on the E. coli infection risk. The highest levels of resistance among all E. coli isolates were observed against ampicillin (88.8%) and amoxicillin/clavulanic acid (62.2%). Among E. coli isolates, 17.7% were classified as multidrug resistant. Escherichia coli isolates showed different pulsotypes and dominant epidemic clones were not detected.
Conclusions
Our data indicate that antibiotic prophylaxis in the presence of symptoms such as chorioamnionitis and premature rupture of membranes did not help reduce the risk of E. coli infection. Multivariate analysis demonstrated only one significant risk factor for E. coli infection among infants with a birth weight <1.5 kg, that is, the impact of the NICU, it means that both neonatal care and care during pregnancy and labour were found to be significant.
Similar content being viewed by others
Background
Infections in newborns, particularly those with very low birth weight of <1.5 kg, remain one of the most significant problems in modern medicine. Escherichia coli is an important cause of neonatal bloodstream and respiratory tract infections and is associated with high mortality [1, 2].
In late-onset infection, E. coli may be acquired from the mother or the environment and may enter the bloodstream from the urinary or gastrointestinal tract [3]. Several studies have demonstrated a high rate of transmission of E. coli within families and households [4, 5]. Escherichia coli species comprise commensal strains, intestinal pathogenic strains and others that cause infections outside the gastrointestinal system, such as extra-intestinal pathogenic E. coli. Extra-intestinal infections due to E. coli are common in all age groups and include bacteremia, urinary tract infection, meningitis (mostly in neonates), nosocomial pneumonia and others [6].
Cephalosporin, fluoroquinolone and trimethoprim-sulfamethoxazole are widely used to treat infections caused by E. coli and resistance to these agents is responsible for treatment failure [7]. The development of multidrug resistance (MDR) among E. coli isolates is also of concern [8].
Enterobacteriaceae-producing extended-spectrum β-lactamases (ESBLs) have emerged as serious pathogens in hospitals and have been increasingly implicated in nosocomial outbreaks in neonatal intensive care units (NICUs). They have important therapeutic implications as they exhibit resistance to several antimicrobial agents, including third-generation cephalosporins, extended-spectrum penicillin and monobactam. Carbapenems and cephamycins represent the only classes of antibiotics active against ESBLs [9].
Escherichia coli has been reported as one of the major causes of neonatal infections that may cause high morbidity and mortality [10]. Potential risk factors of E. coli neonatal infections have also been analysed.
Considering the increasing antimicrobial resistance and mortality in NICUs, there is an urgent need to understand the epidemiology and risk factors of E. coli infections. Therefore, the aim of our study was to investigate the epidemiology of infections caused by E. coli in five Polish NICUs, resistance to antibiotics and potential risk factors that may contribute to such infections (E. coli infections were compared with infections caused by other bacteria). We also considered the possibility of using data on E. coli infection in assessing the quality of perinatal care. All these data may contribute to infection prevention and better control, and the routes of transmission of E. coli were also evaluated.
Methods
Ethics
The Polish Neonatal Surveillance Network (PNSN), whose director is Prof. Ewa Helwich from the Institute of Mother and Child in Warsaw, covered six Neonatal Clinics within the territory of Poland: Clinic of Neonatology and Intensive Neonatal Care, Institute of Mother and Child, Warsaw, Clinic of Neonatology, Jagiellonian University Medical College, Krakow, Clinic of Neonatology and Intensive Neonatal Care, Warsaw Medical University, Warsaw, Clinic of Neonatology, Polish Mothers’ Memorial Hospital-Research Institute, Lodz, Department of Neonatal Diseases, Pomeranian Medical University, Szczecin and Gynaecology and Obstetrics Hospital Medical University Poznan. The Polish Neonatal Surveillance Network was established on 6th of September 2007 (Agreement; see attachment). The Chair of Microbiology cooperated within the program of the PNSN. Utilization of data collected in the PNSN for scientific purposes was approved by the Bioethics Committee of Jagiellonian University Medical College (Chairperson Prof. Piotr Thor) – no. KBET/221/B/2011 ) on 27 October 2011 (Appropoval; see attachement). All data entered into the electronic database and analysed during this study were previously anonymised and de-identified.
Study population
A prospective surveillance of infections was conducted between 1 January 2009 and 31 December 2012 in five from six NICUs that participated in the activities of the PNSN.
The study covered 1,768 newborns. All episodes of infection were subjected to registration, regardless of the time of occurrence of the first symptoms. Case patients were defined according to Gastmeier et al. [11], with modifications for neonates with birth weight <1.5 kg. Early-onset infection (EOI) was defined as infection diagnosed within 3 days of delivery. The occurrence of chorioamnionitis was based on clinical data (all cases) and histopathology examination of the placenta (~65% of cases).
Bacterial isolates
Various diagnostic specimens including blood, and tracheal/bronchial secretions were collected for culture and assessment of the microbial aetiology of infections. Altogether, 96 E. coli strains were isolated, and the present study covered 90 isolates (six were not stored). Isolates were identified by the automated identification system (VITEK 2; bioMérieux, Warsaw, Poland). Isolates from other infections were also analysed. Aetiological factors of other infections were: other Gram-negative bacteria, Gram-positive bacteria, atypical bacteria and yeast.
Antimicrobial susceptibility
All isolates were tested using disk diffusion antimicrobial susceptibility methods on Mueller–Hinton agar plates according to the current EUCAST guidelines (European Committee on Antimicrobial Susceptibility Testing. Clinical breakpoint tables v. 3.1; http://www.eucast.org v.3.1, accessed: 11.02.2013). ESBL activity was detected with a modified double disk synergy test using a combination of cefotaxime, ceftazidime, cefepime and aztreonam discs, placed 20 mm apart around the disc containing amoxicillin/clavulanic acid [12].
Pulsed-field gel electrophoresis (PFGE)
PFGE was used to determine the possible horizontal transfer of E. coli strains among patients. All isolates were analysed using the standardized PFGE protocol developed at the Centers for Disease Control and Prevention by the PulseNet program http://ttp://www.cdc.gov/pulsenet/pathogens/ecoli.html (accessed: 11.02.2013). Genomic DNA was digested with 10 U XbaI (ThermoScientific, ABO, Gdansk, Poland). The resulting DNA fingerprinting was analysed using the CHEF III PFGE system (BioRad, Warsaw, Poland) in 0.5 Tris–borate–EDTA buffer at 14°C at 6 V for 20 h with a ramped pulse time of 2.2–54.2 s. The GelCompar (Applied Maths) was used for cluster analysis using the Dice coefficient and the unweighted pair group method with arithmetic mean.
Risk factors
Several potential risk factors were compared according to infections caused by E. coli and by other strains: birth weight, gestational age, CRIB, Apgar score, sex, type of birth, feeding patterns (breastfeeding, trophic feeding, or parenteral nutrition), perinatal antibiotic prophylaxis, prolonged premature rupture of membranes (PROM).
Statistical analysis
The influence of type of care and sociodemographic characteristics on the epidemiology of E. coli/other infection was analysed with several statistical techniques, depending on type and distribution of analysed variables. The relation between probability of E. coli and continuous parameters (age, length of stay) was based on simple analysis of variance (ANOVA). If the distribution of continuous variables significantly differed from normality, the nonparametric alternative to ANOVA, the Kruskal–Wallis test, was used. For the contingency of nominal characters frequency test: Pearson’s chi-square (χ2) and likelihood ratio was used. The common influence of risk factors on E. coli identification was analysed with a generalised linear model. Because of the categorical character of the effect and combined — numerical as well as categorical — types of the predictors, the model was constructed for binominal distribution of dependent variables and logit-linked function. A p value <0.05 was considered significant. All analyses were performed using JMP version 9.3 (SAS Institute Inc., Cary, NC, USA).
Results
Non-E. coliinfections
Neonates with infections of non-E. coli aetiology were characterised by better general condition at birth (Table 1) than neonates with E. coli infections. Significantly fewer infants had adverse perinatal outcomes, such as PROM or chorioamnionitis, while significantly more newborns were delivered by caesarean section. The fatality rate of newborns with non-E. coli infections was two times lower than in those with E. coli infections.
Escherichia coliinfections
The incidence of E. coli infections was 5.4% and 2.0/1,000 patient-days. EOIs accounted for 12.2% of all infections and late-onset infections (LOIs) accounted for 10.5%. The incidence of EOIs was 1.7% and LOIs was 3.7%. The occurrence of E. coli infections (both EOI and LOI) depended significantly on the NICU (χ2 = 73,836; p < 0.0001) and varied between 3.9% and 17.9%. The most common E. coli infection was pneumonia (53.1%) and bloodstream infection (40.6%). Escherichia coli infections were diagnosed at day 17 on average (median: 12 days).
The mean length of hospitalisation (from birth to discharge, or until a weight of 1.8 kg was reached) of newborns with non-E. coli infections was significantly shorter: 48 versus 55 days (infants with E. coli infection).
The gestational age and birth weight of newborns with E. coli infections were lower than in those without infection or with other infections (Table 1). The other risk factors of E. coli infections were: low Apgar 1 and Apgar 5 scores, total parenteral nutrition, chorioamnionitis, or PROM diagnosed during pregnancy or delivery (Table 1).
Caesarean section reduced the risk of E. coli infection, although perinatal antibiotic prophylaxis (PAP) (no standardised protocol) did not reduce the risk of infection; that is, despite the use of PAP, E. coli infections were observed significantly more frequently (Table 1).
Multivariate analysis
Multivariate analysis that took into account the combined effect of intrapartum antibiotic prophylaxis (IAP), PROM, chorioamnionitis and delivery mode (caesarean section) showed no significant effect on the risk of E. coli infection for factors other than the mode of delivery. However, multivariate analysis that took into account the combined effect of demographic data (sex, gestational age and birth weight) and place of birth (NICU) showed that only the place of hospitalisation had a significant effect on E. coli infection risk.
Finally, a multivariate analysis of maternal influences on IAP, PROM, amniotic inflammation, type of labour, gestational age, NICU and the likelihood of specific E.coli infections (vs. other infections) showed a significant statistical relationship with the complete model.
Escherichia coli resistance and similarity of strains
Analysis of the isolates showed that the highest levels of resistance among all E. coli isolates were observed against ampicillin (88.8%) and amoxicillin/clavulanic acid (62.2%), trimethoprim/sulfamethoxazole (34.4%) and aztreonam (33.3%). The ESBL phenotype was found among 25 isolates (27.7%). Sixteen of the ESBL-positive strains (17.7%) were also reported as resistant to at least two other groups of antibiotics (fluoroquinolones, aminoglycosides or trimethoprim–sulfamethoxazole), which allows us to consider these strains as MDR organisms (Table 2).
Resistance to ampicillin (p = 0.0375), amoxicillin/clavulanic acid, ceftazidime, cefotaxime, cefuroxime, cefepime, aztreonam (all p-values <0.0001), amikacin (p = 0.0012), gentamicin (p = 0.0016), tobramycin (p < 0.0001), trimethoprim/sulfamethoxazole (p = 0.0016) and ciprofloxacin (p < 0.0001) in a group of ESBL-positive strains was significantly higher than in ESBL-negative strains. Only five of the studied isolates studies were resistant to one of four tested carbapenems; three of these isolates belong to non-ESBL strains. Almost all E. coli strains (97.8%) were susceptible to tigecycline and piperacillin/tazobactam.
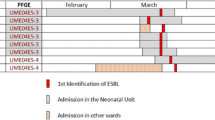
Escherichia coli isolates showed that different pulsotypes and dominant epidemic clones were not detected. Cluster analysis based on PFGE of the 90 isolates showed 71 unique types, some of which were <70% similar, suggesting a genotypically variable population (Figure 1). Isolates that have identical pulsotypes usually were derived from the same patient (as in the case of 11 isolates) or were isolated from different patients of the same NICU in the same period of time (in the case of seven isolates). The location of the NICU and the site of the isolation did not appear to have a correlation in the cluster analysis.
Discussion
Infections in newborns, particularly those with very low and extremely low birth weights of <1.5 kg, are a significant problem. Escherichia coli is responsible for a smaller fraction of neonatal infections than other microorganisms, but it is associated with the highest mortality [13].
Data collected by the European Antimicrobial Resistance Surveillance Network confirm that in Europe from 2002 to 2009 the occurrence of E. coli in bloodstream infections increased by more (71%) than Staphylococcus aureus (34%), which indicates the growing importance of E. coli in the epidemiology of infections [14] and the need for detailed molecular studies of these strains. Our study was performed to characterise the population of E. coli isolated from neonatal infections; mainly from pneumonia and bloodstream infections from Polish NICUs.
According to other data, E. coli causes about 9% of infections [15] or 5–13% of LOI in NICUs [15–17]. Additionally, in Polish NICUs, E. coli was the most frequently isolated pathogen in early-onset PNSN infections [18]. In recent years, E. coli infections in NICUs have become more prevalent, and it is believed that this is mainly owing to the implementation of PAP [19–21].
Other studies on E. coli have identified that low gestational age [22], duration of hospital stay [23] and use of antibiotics [24–26] are the risk factors of colonisation and infection among newborns. Our results support these data, and we hypothesise that another major risk factor may be maternal colonisation. According to Denkel et al., colonised mothers may be an important reservoir of E. coli and significant risk factors for E. coli colonisation, and consequently, E. coli infections of very low birth weight infants [27].
Unfortunately, screening of mothers was not performed in NICUs of PNSN, but our PFGE results support our hypothesis: the population of E. coli was divergent and no epidemic clones were identified. That is totally different than for other species such as methicillin-resistant S. aureus or Klebsiella pneumoniae [28].
The mode of delivery seems to be a protective factor against E. coli infections and also supports the hypothesis on maternal–neonatal transmission. About 17.9% of neonates without infections were born vaginally, compared with 35.4% of those with E. coli infection. The vagina is a significant reservoir of E. coli, which can be critical in vertical transmission of E. coli infection. Our results indicate that perinatal factors and vaginal transmission may also significantly affect the epidemiology of E. coli infection, including LOI, which could be important for control of hospital-acquired infections.
In our study, the fatality rate of E. coli infections was almost twice as high as that of infections of other aetiology. The reason for this was mainly the high pathogenicity of E. coli: the majority of tested strains belonged to one of two virulence groups (B2, 68.9%, or D, 17.8%) [6, 29]. Isolates from the B2 group had significantly more virulence genes than isolates from other groups; in particular, fimH, sfa, ireA, fhuA, fyuA and fepA genes were more likely to occur [29]. Similarly, 36.6% of strains belonged to the ST131 clone and showed a high level of virulence and resistance, which could play an important role in the epidemiological success of this sequence type. Clonal group ST131 contained slightly more than one-third of the studied isolates and had a link to ESBL production [29].
Among ESBL-positive isolates, which accounted for 27.7% of all isolates, higher levels of resistance to aminoglycosides and also ciprofloxacin and trimethoprim-sulfamethoxazole were observed compared with non-ESBL isolates. The higher level of resistance may contribute to the fact that such resistance is localised mainly on plasmids and may be easily transferred not only among E. coli isolates, but also among isolates belonging to other species. Using of one of the antibiotics (β-lactam, aminoglycoside, or fluoroquinolones) may lead to the co-selection of fluoroquinolone resistance by β-lactams or aminoglycosides, and vice versa β-lactams or aminoglycoside resistance by fluoroquinolones [30].
Conclusions
Our study showed that there are some risk factors that should be considered during perinatal care. Routine screening of mothers and newborns for E. coli should be implemented in maternity care to decrease morbidity and mortality. Pre-partum hospitalisation for premature labour should be an indication for screening by rectal swabs, prenatally and on the day of birth. Unfortunately, the presented data indicate that PAP in the presence of symptoms such as chorioamnionitis and PROM did not help to reduce the risk of E. coli infections. Multivariate analysis demonstrated only one significant risk factor for E. coli infection among infants with a birth weight <1.5 kg, namely, the impact of the NICU.
Consent
Electronic database created as the result of continuous prospective targeted surveillance of infections was used in the study.
Participation of hospitals in PNSN was voluntary and confidential. Utilization of data collected in PNSN for the scientific purpose was approved by the Bioethics Committee of Jagiellonian University Medical College – no. KBET/221/B/2011. All data entered into the electronic database and analyzed during preparing this article were previously anonymized and de-identified. Those data were obtained under routine treatment and diagnostic procedures performed during patients’ hospitalization. No additional samples were collected for testing. According to Polish law, utilization this kind of data for scientific purpose does not demand patients’ agreement or even information that data are collected in the database.
Abbreviations
- BSI:
-
Bloodstream infection
- EOI:
-
Early-onset infection
- ESBL:
-
Extended-spectrum β-lactamase
- IAP:
-
Intrapartum antibiotic prophylaxis
- LOI:
-
Late-onset infection
- MDR:
-
Multidrug resistance
- NICU:
-
Neonatal intensive care unit
- PAP:
-
Perinatal antibiotic prophylaxis
- PFGE:
-
Pulsed-field gel electrophoresis
- PROM:
-
Premature rupture of membranes.
References
Stoll BJ, Hansen N, Higgins RD, Fanaroff AA, Duara S, Goldberg R, Laptook A, Walsh M, Oh W, Hale E: National institute of child health and human development: very low birth weight preterm infants with early onset neonatal sepsis: the predominance of gram-negative infections continues in the national institute of child health and human development neonatal research network, 2002–2003. Pediatr Infect Dis J. 2005, 24: 635-639. 10.1097/01.inf.0000168749.82105.64.
Doit C, Mariani-Kurkdjian P, Mahjoub-Messai F, Bidet P, Bonacorsi S, Carol A, Varon E, Bingen E: Epidemiology of paediatric community-acquired bloodstream infections in a children’s hospital in Paris, France, 2001 to 2008. Diagn Microbiol Infect Dis. 2010, 66: 332-335. 10.1016/j.diagmicrobio.2009.10.012.
Soto SM, Bosch J, Jimenez De Anta MT, Vila J: Comparative study of virulence traits of Escherichia coli clinical isolates causing early and late neonatal sepsis. J Clin Microbiol. 2008, 46: 1123-1125. 10.1128/JCM.01682-07.
Birgy A, Mariani-Kurkdjian P, Bidet P, Doit C, Genel N, Courroux C, Arlet G, Bingen E: Characterization of extended-spectrum-beta-lactamase-producing Escherichia coli strains involved in maternal-fetal colonization: prevalence of E. coli ST131. J Clin Microbiol. 2013, 51 (6): 1727-1732. 10.1128/JCM.03255-12.
Rodríguez-Baño J, López-Cerero L, Navarro MD, Díaz De Alba P, Pascual A: Faecal carriage of extended-spectrum β-lactamase-producing Escherichia coli: prevalence, risk factors and molecular epidemiology. J Antimicrob Chemother. 2008, 62: 1142-1149. 10.1093/jac/dkn293.
Pitout JD: Extraintestinal: pathogenic Escherichia coli: a combination of virulence with antibiotic resistance. Front Microbiol. 2012, 3: 9-
Woodford N, Turton JF, Livermore DM: Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011, 35: 736-755. 10.1111/j.1574-6976.2011.00268.x.
Hawkey PM, Jones AM: The changing epidemiology of resistance. J Antimicrob Chemother. 2009, 64: 3-10. 10.1093/jac/dkp256.
Coque TM, Baquero F, Canton R: Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill. 2008, 13: 47-
Stoll BJ, Hansen N, Fanaroff AA, Wright LL, Carlo WA, Ehrenkranz RA, Lemons JA, Donovan EF, Stark AR, Tyson JE, Oh W, Bauer CR, Korones SB, Shankaran S, Laptook AR, Stevenson DK, Papile LA, Poole WK: Changes in pathogens causing early-onset sepsis in very-low-birth-weight infants. N Engl J Med. 2002, 25 (4): 240-247.
Gastmeier P, Geffers C, Schwab F, Mitzner J, Oblader M, Ruden H: Development of a surveillance system for nosocomial infections: the component for neonatal intensive care in Germany. J Hosp Infect. 2004, 57: 126-131. 10.1016/j.jhin.2003.12.038.
Drieux L, Brossier F, Sougakoff W, Jarlier V: Phenotypic detection of extended-spectrum β-lactamase production in Enterobacteriaceae: review and bench guide. Clin Microbiol Infect. 2008, 14: 90-103. 10.1111/j.1469-0691.2007.01846.x.
Stoll BJ, Hansen N, Fanaroff AA, Wright LL, Carlo WA, Ehrenkranz RA, Lemons JA, Donovan EF, Stark AR, Tyson JE, Oh W, Bauer CR, Korones SB, Shankaran S, Laptook AR, Stevenson DK, Papile LA, Poole WK: Late-onset sepsis in very low birth weight neonates: the experience of the NICHD Neonatal Research Network. Pediatrics. 2002, 110: 285-291. 10.1542/peds.110.2.285.
Gagliotti C, Balode A, Baquero F, Degener J, Grundmann H, Gür D, Jarlier V, Kahlmeter G, Monen J, Monnet DL, Rossolini GM, Suetens C, Weist K, Heuer O: EARS-Net Participants (Disease Specific Contact Points for AMR). Escherichia coli and Staphylococcus aureus: bad news and good news from the European Antimicrobial Resistance Surveillance Network (EARS-Net, formerly EARSS), 2002 to 2009. Euro Surveill. 2011, 17: 19819-Erratum in: Euro Surveillance 2011, 16:19834
Babazono A, Kitajima H, Nishimaki S, Nakamura T, Shiga S, Hayakawa M, Tanaka T, Sato K, Nakayama H, Ibara S, Une H, Doi H: Risk factors for nosocomial infections in the neonatal intensive care unit by the Japanese National Infection Surveillance (JANIS). Acta Med Okayama. 2008, 62 (4): 261-26.
Vergnano S, Menson E, Kennea N, Embleton N, Russell AB, Watts T, Robinson MJ, Collinson A, Heath PT: Neonatal infections in England: the NeonIT surveillance network. Arch Dis Child Fetal Neonatal Ed. 2011, 96: F9-F1. 10.1136/adc.2009.178798.
Geffers C, Bearwolff S, Schwab F, Gastmeier P: Incidence of healthcare-associated infections in high-risk neonates: results from the German surveiilance system for very-low-birthweight infants. J Hosp Infect. 2008, 68: 214-221. 10.1016/j.jhin.2008.01.016.
Wójkowska-Mach J, Borszewska-Kornacka M, Domańska J, Gadzinowski J, Gulczyńska E, Helwich E, Kordek A, Pawlik D, Szczapa J, Klamka J, Heczko PB: Early-onset Infections of Very-low-birth-weight Infants in Polish Neonatal Intensive Care Units. Pediatr Infect Dis J. 2012, 31 (7): 691-695. 10.1097/INF.0b013e3182567b74.
Gaynes RP, Edwards JR, Jarvis WR, Culver DH, Tolson JS, Martone WJ: Nosocomial infections among neonates in high-risk nurseries in the United States. National Nosocomial Infections Surveillance System. Pediatrics. 1996, 98: 357-361.
Lin CY, Hsu CH, Huang FY, Chang JH, Hung HY, Kao HA, Peng CC, Jim WT, Chi H, Chiu NC, Chang TY, Chen CY, Chen CP: The changing face of early-onset neonatal sepsis after the implementation of a maternal group B Streptococcus screening and intrapartum prophylaxis policy–a study in one medical center. Pediatr Neonatol. 2011, 52 (2): 78-84. 10.1016/j.pedneo.2011.02.001.
Shane AL, Stoll BJ: Recent developments and current issues in the epidemiology, diagnosis, and management of bacterial and fungal neonatal sepsis. Am J Perinatol. 2013, 30 (2): 131-141. 10.1055/s-0032-1333413.
Millar M, Philpott A, Wilks M, Whiley A, Warwick S, Hennessy E, Coen P, Kempley S, Stacey F, Costeloe K: Colonization and persistence of antibiotic-resistant Enterobacteriaceae strains in infants nursed in two neonatal intensive care units in East London, United Kingdom. J Clin Microbiol. 2008, 46: 560-567. 10.1128/JCM.00832-07.
Shakil S, Akram M, Ali SM, Khan AU: Acquisition of extended-spectrum β-lactamase producing Escherichia coli strains in male and female infants admitted to a neonatal intensive care unit: molecular epidemiology and analysis of risk factors. J Med Microbiol. 2010, 59: 948-954. 10.1099/jmm.0.020214-0.
Huang Y, Zhuang S, Du M: Risk factors of nosocomial infection with extended-spectrum β-lactamase-producing bacteria in a neonatal intensive care unit in China. Infection. 2007, 35: 339-345. 10.1007/s15010-007-6356-9.
Bromiker R, Ernest N, Meir MB, Kaplan M, Hammerman C, Schimmel MS, Schlesinger Y: Correlation of bacterial type and antibiotic sensitivity with maternal antibiotic exposure in early-onset neonatal sepsis. Neonatol. 2013, 103: 48-53. 10.1159/000342215.
Benenson S, Levin PD, Block C, Adler A, Ergaz Z, Peleg O, Minster N, Gross I, Schaffer K, Moses AE, Cohen MJ: Continuous surveillance to reduce extended-spectrum β-lactamase Klebsiella pneumoniae colonization in the neonatal intensive care unit. Neonatol. 2013, 103: 155-160. 10.1159/000343150.
Denkel LA, Schwab F, Kola A, Leistner R, Garten L, von Weizsäcker K, Geffers C, Gastmeier P, Piening B: The mother as most important risk factor for colonization of very low birth weight (VLBW) infants with extended-spectrum β-lactamase-producing Enterobacteriaceae (ESBL-E). J Antimicrob Chemother. 2014, 69 (8): 2230-2237. 10.1093/jac/dku097.
Gastmeier P, Loui A, Stamm-Balderjahn S, Hansen S, Zuschneid I, Sohr D, Behnke M, Obladen M, Vonberg RP, Rüden H: Outbreaks in neonatal intensive care units - they are not like others. Am J Infect Control. 2007, 35 (3): 172-176. 10.1016/j.ajic.2006.07.007.
Chmielarczyk A, Pobiega M, Wojkowska-Mach J, Romaniszyn D, Adamski P, Heczko PB, Bulanda M: Molecular epidemiology, plasmid analysis, virulence and resistance of Escherichia coli isolated from neonatal intensive care units in Poland. Diagn Microbiol Infect Dis. 2013, 76 (4): 542-545. 10.1016/j.diagmicrobio.2013.04.016.
Bailey JK, Pinyon JL, Anantham S, Hall RM: Commensal Escherichia coli of healthy humans: A reservoir for antibiotic-resistance determinants. J Med Microbiol. 2010, 59 (Pt 11): 1331-1339.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2431/14/274/prepub
Acknowledgements
The authors wish to thank the staff of the NICUs for their help and interest in the study.
Funding
This study was partially supported by a grant from the Ministry of Science and Higher Education (DEC-2011/01/D/N27/00104).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ACH carried out the molecular genetic studies (PFGE), drafted the manuscript and financially supported the study. JWM designed the study, analysed and interpreted the epidemiological data, and drafted the manuscript. DR carried out the antimicrobial susceptibility studies. MP performed the statistical analysis and drafted the manuscript. PA performed the statistical analysis. RL, MBK, EG and AK collected and analysed the data and participated in study design. EH and PBH conceived the study, participated in its design and helped to draft the manuscript. All authors read and approved the final manuscript for publication.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Chmielarczyk, A., Wójkowska-Mach, J., Romaniszyn, D. et al. Mode of delivery and other risk factors for Escherichia coli infections in very low birth weight infants. BMC Pediatr 14, 274 (2014). https://doi.org/10.1186/1471-2431-14-274
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2431-14-274