Abstract
Micro-RNAs (miRs) are important regulators of mRNA and protein expression; the ability of miR expression profilings to distinguish different cancer types and classify their sub-types has been well-described. They also represent a novel biological entity with potential value as tumour biomarkers, which can improve diagnosis, prognosis, and monitoring of treatment response for human cancers. This endeavour has been greatly facilitated by the stability of miRs in formalin-fixed paraffin-embedded (FFPE) tissues, and their detection in circulation. This review will summarize some of the key dysregulated miRs described to date in human epithelial malignancies, and their potential value as molecular bio-markers in FFPE tissues and blood samples. There remain many challenges in this domain, however, with the evolution of different platforms, the complexities of normalizing miR profiling data, and the importance of evaluating sufficiently-powered training and validation cohorts. Nonetheless, well-conducted miR profiling studies should contribute important insights into the molecular aberrations driving human cancer development and progression.
Similar content being viewed by others
Introduction
Micro-RNAs (miRs) are important regulators of mRNA and protein expression which play important yet complex roles in human cancers [1]. Their biogenesis and biological networks are complex (Figure 1); they are first synthesized as large RNA precursors, processed in the nucleus into approximately 70 nt pre-miRs, folded into imperfect stem-loop structures, transported to the cytoplasm, whereupon they are incorporated into RISC (RNA-induced silencing complex) (reviewed in [2]). Cleavage by Argonaute-2, then Dicer, results in an approximately 22-nt mature miR duplex; the "guide" strand is retained within the RISC; the "passenger" strand is degraded. Through the seed region (nt 2 to 8), the miR can then bind to the 3'UTR of target mRNA sequences, preventing protein translation, leading to mRNA degradation. More recently, miRs have also been described to target 5'UTR, and even coding regions of transcripts [3]. The current miRDatabase (http://www.mirbase.org) has catalogued more than 1,300 human sequences. Given their ability to target mRNA with imperfect complementarity, and predicted to regulate the expression of approximately one-third of all human transcripts [4], miRs are considered to be among the largest class of gene regulators [5, 6].
Micro-RNA biogenesis. MiR's are synthesized initially as large RNA precursors (pri-miRs), processed in the nucleus by RNAse III Drosha, and DGCR8 into approximately 70 nt pre-miR, which are transported to the cytoplasm by exportin-5, with subsequent cleavage by another RNAse III enzyme Dicer, with its co-factor TRBP, releasing the approximately 22-nt mature dsmiR. MiR's can negatively regulate their targets in one of two major ways, depending on the degree of complementarity to its target. First, and probably most commonly, one strand of this duplex is incorporated into the RNA-induced silencing complex (RISC), then binds with imperfect complementarity to the 3'-UTR (untranslated region) of mRNA targets, preventing protein translation. Alternatively, miRs can bind with perfect complementarity to the ORF (open reading frame) of target mRNA's with subsequent degradation. Recent evidence also indicates miRs can also bind to either promoters, or coding regions of mRNAs as additional mechanisms of regulation.
Multiple mechanisms can mediate miR dysregulations in human cancers, including chromosomal gains or losses [7], mutations of miR located loci [8], or epigenetic aberrations [8]. Any misstep in miR biogenesis (Figure 1) can also affect miR expression [9, 10], exemplified by the down-regulation of Drosha and Dicer being associated with worse survival in ovarian, lung, and breast cancers [11]. MiRs can be either over- or under-expressed, functioning as tumour suppressors or oncogenes, depending on their downstream target genes [12]. MiR-15a and miR-16-1 are two of the first described down-regulated miRs in chronic lymphocytic leukemia [13], both target Bcl-2 [14]; thus their absence inhibits apoptosis. Alternatively, miR-21, one of the most commonly over-expressed miRs in solid malignancies, targets PTEN [15] and pro-apoptotic genes [16, 17]; hence pro-survival signals dominate.
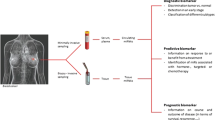
Micro-RNA as bio-markers in epithelial cancers
Biomarkers are biological indicators of disease states, utilized to define tumor subtypes, or assess efficacy of interventions [18]. Useful biomarkers can provide insights into tumorigenesis, and facilitate the development of improved therapies. Some current bio-markers include prostate-specific antigen (PSA) [19], carcinoembryonic antigen (CEA) [20], CA125 [21], and α-fetoprotein [22, 23]. More recently, the role of mRNA or miRs as cancer biomarkers have also been investigated and developed. The prototype mRNA signature is Oncotype DX, the 21-gene set utilized to predict recurrence risks for patients with breast cancer [24].
MiR expression profilings could distinguish different cancer types [12], classify sub-types of prostate or breast cancers [25], identify the tissue origin of tumors [26], and facilitate the diagnosis of colon [27], or lung cancers [28]. MiRs can also predict outcome, such as let-7a [28] and miR-155 [29] for lung cancer, and select patients for targeted therapy (for example, breast cancer [30]). Finally, predictive miR signatures have been reported for several malignancies, such as lung [31–34], hepatocellular [35], esophageal [36], gastric [37], prostate [38] cervical [39], and colon cancers [40].
Micro-RNAs in FFPE samples
The ability to examine FFPEspecimens, a universally standard histologic processing procedure, allows the expeditious discovery and evaluation of potential biomarkers, given their possible link to clinical databases with mature follow-up. Transcript (mRNA) profiling is technically challenging with FFPE samples due to significant RNA degradation during formalin fixation [41, 42], and continued deterioration with storage over time [43]. In contrast, miRs are not significantly affected by fixation, and can be readily extracted from FFPE samples due to their small sizes (approximately 22 nt in length) and remarkable stability [44, 45]. Hence, this greatly enhances the ability to evaluate miRs as cancer biomarkers, leading to a multitude of reports describing miR expressions in many epithelial malignancies, summarized as per anatomical site in Table 1.
As already mentioned, miR-21 up-regulation is the most commonly observed aberrant miR in human cancers, with oncogenic consequences [46] (Table 1). It was first reported in glioblastoma [16], but also described for epithelial cancers such as head and neck, breast, colon, lung, prostate, and others [12, 44, 47]; often associated with worse outcome [40]. Over-expression of miR-21 has been shown to increase cell proliferation, migration, invasion and survival [48]; in contrast, suppression of miR-21 induced apoptosis and decreased cell proliferation and invasion [49].
Mir-155 is another commonly dysregulated miR, wherein the majority of studies report its over-expression associated with tumorigenesis in lymphomas, breast, lung, colon, pancreatic cancers, and others [50]. Aside from these two miRs, there is usually minimal overlap of dysregulated miRs described amongt different studies, even when examining the same cancer type; the same variation as previously observed for mRNA profiling. Perhaps this might relate to multiple redundant "wirings" in human cancers [51], wherein just as four distinct mRNA profiles can all predict for breast cancer relapse [52], a similar phenomenon might also apply to miR profiles, although this remains to be definitively proven.
Micro-RNAs in blood samples
There is emerging interest in the investigation of miRs as non-invasive biomarkers in circulating blood. This was first described in B-cell lymphoma, reporting elevated levels of miR-155, -210 and -21 in patients' sera, with miR-21 associating with relapse-free survival [53]. In epithelial cancers, Mitchell et al. first identified tumor-derived miRs in plasma samples, and suggested that variations in miR abundance reflected tumor burden [54]. MiRs have been detected as free miRs in either plasma or serum, or contained within microvesicles such as exosomes; the latter being minute, natural membrane vesicles secreted by a variety of different cell types [55]. In addition to miRs, exosomes also carry intact and functional mRNA [56], with the probable purpose of transferring information and signals throughout the body [55]. Association of epithelial cancer and exosome miRs was first illustrated in ovarian cancer, wherein tumor-derived miR profiles strongly correlated with levels of peripheral blood-derived exosomal miRs [57]. Similar observations have also been reported for lung cancer [58, 59].
As shown in Table 2, the list of potential blood miR biomarkers is even more diverse than those from tissue studies (Table 1). The greatest degree of overlap was reported for miR-21, miR-196a and miR-210 from four different pancreatic cancer studies [60–63]. As observed for the tissue studies, miR-21 and miR-155 are also the two most common aberrant miRs in circulation with putative diagnostic and prognostic value (Table 2). However, down-regulation of miR-155 was reported in one serum study of ovarian cancer [64]. There is some controversy surrounding miR-155; the majority of reports suggest an oncogenic role; however, in a lung cancer study, its up-regulation predicted for worse outcome for adenocarcinomas, but improved outcome for squamous cell carcinoma patients [65]. One possible tumor suppression function for miR-155 was demonstrated in miR-155 deficient mice, which appeared to reduce oncogenic translocations generated by activation-induced cytidine deaminase (AICD) [66]. Micro-RNA expression levels in circulation can also relate to hormone receptor status in that estrogen negative breast cancer sera samples had higher levels of miR-21 and miR-10b [67]; in contrast. miR-155 was detected for progesterone receptor positive patients [68].
In summary, there are multitudes of reports describing the potential value of miRs as both diagnostic and prognostic bio-markers for human malignancies. None to date, however, have been translated into clinical practice, likely a reflection of its complex biology, and lack of validation studies using appropriately-powered sample sizes.
Challenges of Micro-RNA as bio-markers
Despite the promising data supporting the potential value of miRs as biomarkers, many challenges remain. First, robust platforms, as well as appropriate statistical and bio-computational analyses must be utilized in order to identify potential candidate miR signatures for predicting outcome. Furthermore, such candidate signatures must be validated using independent cohorts statistically powered to confirm the existence of a predictive signature. Second, the selection of the appropriate reference controls is extremely important for normalization of biological variation. Recent reports have observed that some of the commonly-utilized reference miRs, such as RNU43, RNU44 or RNU48, in fact fluctuate with the biological entity of interest [69]; hence it is critical to determine the most stable miRs for each condition under examination. Third, it is conceivable that given the "upstream" effects of miRs, and their biological complexities which we are just starting to unravel, their pattern of expression might be too subtle and variable to serve as robust predictive signatures. Nonetheless, pursuit of investigations such as prognostic signatures, or their measurements in sera/plasma are definitely warranted, particularly when using appropriately-sized population cohorts.
Conclusion
Application of the potential role of miRs as molecular bio-markers in human epithelial malignancies is widely supported by the large number of studies conducted in different cancers. There is great promise that they will aid in the early diagnosis of cancer, and the development of personalized therapies. Further research into miR biogenesis and regulation, along with functional target identifications will definitely lead to an improved understanding of the complex mechanisms underlying human cancer development and progression.
Abbreviations
- AICD:
-
activation-induced cytidine deaminase
- CEA:
-
carcinoembryonic antigen
- FFPE:
-
formalin fixed and paraffin embedded
- miRs:
-
micro-RNAs
- PSA:
-
prostate-specific antigen
- RISC:
-
RNA-induced silencing complex.
References
Garzon R, Calin GA, Croce CM: MicroRNAs in cancer. Annu Rev Med. 2009, 60: 167-179. 10.1146/annurev.med.59.053006.104707.
Kai ZS, Pasquinelli AE: MicroRNA assassins: factors that regulate the disappearance of miRNAs. Nat Struct Mol Biol. 2010, 17: 5-10. 10.1038/nsmb.1762.
Breving K, Esquela-Kerscher A: The complexities of microRNA regulation: Mirandering around the rules. Int J Biochem Cell Biol. 2009, 42: 1316-1329.
Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ: miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34: D140-144. 10.1093/nar/gkj112.
Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z: Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005, 37: 766-770. 10.1038/ng1590.
Stefani G, Slack FJ: Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008, 9: 219-230. 10.1038/nrm2347.
Calin GA Croce CM: MicroRNA signatures in human cancers. Nat Rev Cancer. 2006, 6: 857-866. 10.1038/nrc1997.
Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM: A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005, 353: 1793-1801. 10.1056/NEJMoa050995.
Wiemer EA: The role of microRNAs in cancer: no small matter. Eur J Cancer. 2007, 43: 1529-1544. 10.1016/j.ejca.2007.04.002.
Zhang B, Pan X, Cobb GP, Anderson TA: microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007, 302: 1-12. 10.1016/j.ydbio.2006.08.028.
Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick AM, Deavers MT, Mourad-Zeidan A, Wang H, Mueller P, Lenburg ME, Gray JW, Mok S, Birrer MJ, Lopez-Berestein G, Coleman RL, Bar-Eli M, Sood AK: Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008, 359: 2641-2650. 10.1056/NEJMoa0803785.
Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM: A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006, 103: 2257-2261. 10.1073/pnas.0510565103.
Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM: Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002, 99: 15524-15529. 10.1073/pnas.242606799.
Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM: miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005, 102: 13944-13949. 10.1073/pnas.0506654102.
Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T: MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007, 133: 647-658. 10.1053/j.gastro.2007.05.022.
Chan JA, Krichevsky AM, Kosik KS: MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005, 65: 6029-6033. 10.1158/0008-5472.CAN-05-0137.
Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY: miR-21-mediated tumor growth. Oncogene. 2007, 26: 2799-2803. 10.1038/sj.onc.1210083.
Ratain MJ, Glassman RH: Biomarkers in phase I oncology trials: signal, noise, or expensive distraction?. Clin Cancer Res. 2007, 13: 6545-6548. 10.1158/1078-0432.CCR-07-2133.
Balk SP, Ko YJ, Bubley GJ: Biology of prostate-specific antigen. J Clin Oncol. 2003, 21: 383-391. 10.1200/JCO.2003.02.083.
Thomas SN, Tong Z, Stebe KJ, Konstantopoulos K: Identification, characterization and utilization of tumor cell selectin ligands in the design of colon cancer diagnostics. Biorheology. 2009, 46: 207-225.
Osman N, O'Leary N, Mulcahy E, Barrett N, Wallis F, Hickey K, Gupta R: Correlation of serum CA125 with stage, grade and survival of patients with epithelial ovarian cancer at a single centre. Ir Med J. 2008, 101: 245-247.
Heyward WL, Lanier AP, Bender TR, McMahon BJ, Kilkenny S, Paprocki TR, Kline KT, Silimperi DR, Maynard JE: Early detection of primary hepatocellular carcinoma by screening for alpha-fetoprotein in high-risk families. A case-report. Lancet. 1983, 2: 1161-1162.
Lange PH, Vogelzang NJ, Goldman A, Kennedy BJ, Fraley EE: Marker half-life analysis as a prognostic tool in testicular cancer. J Urol. 1982, 128: 708-711.
Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N: A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004, 351: 2817-2826. 10.1056/NEJMoa041588.
Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, Fedele V, Ginzinger D, Getts R, Haqq C: Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006, 5: 24-10.1186/1476-4598-5-24.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR: MicroRNA expression profiles classify human cancers. Nature. 2005, 435: 834-838. 10.1038/nature03702.
Michael MZ, O'Connor SM, van Holst Pellekaan NG, Young GP, James RJ: Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003, 1: 882-891.
Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T: Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004, 64: 3753-3756. 10.1158/0008-5472.CAN-04-0637.
Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T: A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005, 65: 9628-9632. 10.1158/0008-5472.CAN-05-2352.
Chen JQ, Russo J: ERalpha-negative and triple negative breast cancer: molecular features and potential therapeutic approaches. Biochim Biophys Acta. 2009, 1796: 162-175.
Du L, Schageman JJ, Irnov , Girard L, Hammond SM, Minna JD, Gazdar AF, Pertsemlidis A: MicroRNA expression distinguishes SCLC from NSCLC lung tumor cells and suggests a possible pathological relationship between SCLCs and NSCLCs. J Exp Clin Cancer Res. 2010, 29: 75-10.1186/1756-9966-29-75.
Raponi M, Dossey L, Jatkoe T, Wu X, Chen G, Fan H, Beer DG: MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009, 69: 5776-5783. 10.1158/0008-5472.CAN-09-0587.
Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC: Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006, 9: 189-198. 10.1016/j.ccr.2006.01.025.
Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, Cheng CL, Yu CJ, Lee YC, Chen HS, Su TJ, Chiang CC, Li HN, Hong QS, Su HY, Chen CC, Chen WJ, Liu CC, Chan WK, Chen WJ, Li KC, Chen JJ, Yang PC: MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008, 13: 48-57. 10.1016/j.ccr.2007.12.008.
Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM, Tang ZY, Wang XW: Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008, 47: 897-907. 10.1002/hep.22160.
Guo Y, Chen Z, Zhang L, Zhou F, Shi S, Feng X, Li B, Meng X, Ma X, Luo M, Shao K, Li N, Qiu B, Mitchelson K, Cheng J, He J: Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res. 2008, 68: 26-33. 10.1158/0008-5472.CAN-06-4418.
Li X, Zhang Y, Ding J, Wu K, Fan D: Survival prediction of gastric cancer by a seven-microRNA signature. Gut. 2010, 59: 579-585. 10.1136/gut.2008.175497.
Tong AW, Fulgham P, Jay C, Chen P, Khalil I, Liu S, Senzer N, Eklund AC, Han J, Nemunaitis J: MicroRNA profile analysis of human prostate cancers. Cancer Gene Ther. 2009, 16: 206-216.
Hu X, Schwarz JK, Lewis JS, Huettner PC, Rader JS, Deasy JO, Grigsby PW, Wang X: A microRNA expression signature for cervical cancer prognosis. Cancer Res. 2010, 70: 1441-1448. 10.1158/0008-5472.CAN-09-3289.
Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, Croce CM, Harris CC: MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008, 299: 425-436. 10.1001/jama.299.4.425.
Bresters D, Schipper ME, Reesink HW, Boeser-Nunnink BD, Cuypers HT: The duration of fixation influences the yield of HCV cDNA-PCR products from formalin-fixed, paraffin-embedded liver tissue. J Virol Methods. 1994, 48: 267-272. 10.1016/0166-0934(94)90125-2.
Macabeo-Ong M, Ginzinger DG, Dekker N, McMillan A, Regezi JA, Wong DT, Jordan RC: Effect of duration of fixation on quantitative reverse transcription polymerase chain reaction analyses. Mod Pathol. 2002, 15: 979-987. 10.1097/01.MP.0000026054.62220.FC.
Cronin M, Pho M, Dutta D, Stephans JC, Shak S, Kiefer MC, Esteban JM, Baker JB: Measurement of gene expression in archival paraffin-embedded tissues: development and performance of a 92-gene reverse transcriptase-polymerase chain reaction assay. Am J Pathol. 2004, 164: 35-42. 10.1016/S0002-9440(10)63093-3.
Hui AB, Shi W, Boutros PC, Miller N, Pintilie M, Fyles T, McCready D, Wong D, Gerster K, Waldron L, Jurisica I, Penn LZ, Liu FF: Robust global micro-RNA profiling with formalin-fixed paraffin-embedded breast cancer tissues. Lab Invest. 2009, 89: 597-606. 10.1038/labinvest.2009.12.
Nelson PT, Baldwin DA, Scearce LM, Oberholtzer JC, Tobias JW, Mourelatos Z: Microarray-based, high-throughput gene expression profiling of microRNAs. Nat Methods. 2004, 1: 155-161. 10.1038/nmeth717.
Krichevsky AM, Gabriely G: miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009, 13: 39-53.
Hui AB, Lenarduzzi M, Krushel T, Waldron L, Pintilie M, Shi W, Perez-Ordonez B, Jurisica I, O'Sullivan B, Waldron J, Gullane P, Cummings B, Liu FF: Comprehensive MicroRNA profiling for head and neck squamous cell carcinomas. Clin Cancer Res. 2010, 16: 1129-1139. 10.1158/1078-0432.CCR-09-2166.
Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, Li Y: MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008, 27: 4373-4379. 10.1038/onc.2008.72.
Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H: MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008, 27: 2128-2136. 10.1038/sj.onc.1210856.
Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, Kroesen BJ, van den Berg A: BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005, 207: 243-249. 10.1002/path.1825.
Hanahan D, Weinberg RA: Hallmarks of cancer: the next generation. Cell. 2011, 144: 646-674. 10.1016/j.cell.2011.02.013.
Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DS, Nobel AB, van't Veer LJ, Perou CM: Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006, 355: 560-569. 10.1056/NEJMoa052933.
Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, Hatton CS, Harris AL: Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008, 141: 672-675. 10.1111/j.1365-2141.2008.07077.x.
Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M: Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008, 105: 10513-10518. 10.1073/pnas.0804549105.
Cocucci E, Racchetti G, Rupnik M, Meldolesi J: The regulated exocytosis of enlargeosomes is mediated by a SNARE machinery that includes VAMP4. J Cell Sci. 2008, 121: 2983-2991. 10.1242/jcs.032029.
Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO: Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007, 9: 654-659. 10.1038/ncb1596.
Taylor DD, Gercel-Taylor C: MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008, 110: 13-21. 10.1016/j.ygyno.2008.04.033.
Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH: Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009, 10: 42-46. 10.3816/CLC.2009.n.006.
Silva J, García V, Zaballos Á, Provencio M, Lombardía L, Almonacid L, García JM, Dominguez G, Peña C, Diaz R, Herrera M, Varela A, Bonilla F: Vesicle-related microRNAs in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur Respir J. 2011, 37: 617-623. 10.1183/09031936.00029610.
Ali S, Almhanna K, Chen W, Philip PA, Sarkar FH: Differentially expressed miRNAs in the plasma may provide a molecular signature for aggressive pancreatic cancer. Am J Transl Res. 2011, 3: 28-47.
Kong X, Du Y, Wang G, Gao J, Gong Y, Li L, Zhang Z, Zhu J, Jing Q, Qin Y, Li Z: Detection of differentially expressed microRNAs in serum of pancreatic ductal adenocarcinoma patients: miR-196a could be a potential marker for poor prognosis. Dig Dis Sci. 2011, 56: 602-609. 10.1007/s10620-010-1285-3.
Ho AS, Huang X, Cao H, Christman-Skieller C, Bennewith K, Le QT, Koong AC: Circulating miR-210 as a Novel Hypoxia Marker in Pancreatic Cancer. Transl Oncol. 2010, 3: 109-113.
Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, Frazier ML, Killary AM, Sen S: MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila). 2009, 2: 807-813. 10.1158/1940-6207.CAPR-09-0094.
Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, Cohn DE: The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. 2009, 112: 55-59. 10.1016/j.ygyno.2008.08.036.
Donnem T, Eklo K, Berg T, Sorbye SW, Lonvik K, Al-Saad S, Al-Shibli K, Andersen S, Stenvold H, Bremnes RM, Busund LT: Prognostic impact of MiR-155 in non-small cell lung cancer evaluated by in situ hybridization. J Transl Med. 2011, 9: 6-10.1186/1479-5876-9-6.
Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai TH, Robbiani DF, Di Virgilio M, Reina San-Martin B, Heidkamp G, Schwickert TA, Eisenreich T, Rajewsky K, Nussenzweig MC: MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008, 28: 630-638. 10.1016/j.immuni.2008.04.002.
Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ: Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010, 251: 499-505. 10.1097/SLA.0b013e3181cc939f.
Zhu W, Qin W, Atasoy U, Sauter ER: Circulating microRNAs in breast cancer and healthy subjects. BMC Res Notes. 2009, 2: 89-10.1186/1756-0500-2-89.
Gee HE, Buffa FM, Camps C, Ramachandran A, Leek R, Taylor M, Patil M, Sheldon H, Betts G, Homer J, West C, Raqoussis J, Harris AL: The small-nucleolar RNAs commonly used for microRNA normalisation correlate with tumour pathology and prognosis. Br J Cancer. 104: 1168-1177.
Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, Wells W, Kauppinen S, Cole CN: Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007, 67: 11612-11620. 10.1158/0008-5472.CAN-07-5019.
Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY: MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008, 14: 2348-2360. 10.1261/rna.1034808.
Lebanony D, Benjamin H, Gilad S, Ezagouri M, Dov A, Ashkenazi K, Gefen N, Izraeli S, Rechavi G, Pass H, Nonaka D, Li J, Spector Y, Rosenfeld N, Chajut A, Cohen D, Aharonov R, Mansukhani M: Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J Clin Oncol. 2009, 27: 2030-2037. 10.1200/JCO.2008.19.4134.
Navarro A, Diaz T, Gallardo E, Vinolas N, Marrades RM, Gel B, Campayo M, Quera A, Bandres E, Garcia-Foncillas J, Ramirez J, Monzo M: Prognostic implications of miR-16 expression levels in resected non-small-cell lung cancer. J Surg Oncol. 2010, 103: 411-415.
Xiao B, Guo J, Miao Y, Jiang Z, Huan R, Zhang Y, Li D, Zhong J: Detection of miR-106a in gastric carcinoma and its clinical significance. Clin Chim Acta. 2009, 400: 97-102. 10.1016/j.cca.2008.10.021.
Zhang Y, Guo J, Li D, Xiao B, Miao Y, Jiang Z, Zhuo H: Down-regulation of miR-31 expression in gastric cancer tissues and its clinical significance. Med Oncol. 2009, 27: 685-689.
Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM: MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007, 297: 1901-1908. 10.1001/jama.297.17.1901.
Habbe N, Koorstra JB, Mendell JT, Offerhaus GJ, Ryu JK, Feldmann G, Mullendore ME, Goggins MG, Hong SM, Maitra A: MicroRNA miR-155 is a biomarker of early pancreatic neoplasia. Cancer Biol Ther. 2009, 8: 340-346. 10.4161/cbt.8.4.7338.
du Rieu MC, Torrisani J, Selves J, Al Saati T, Souque A, Dufresne M, Tsongalis GJ, Suriawinata AA, Carrere N, Buscail L, Cordelier P: MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions. Clin Chem. 2010, 56: 603-612. 10.1373/clinchem.2009.137364.
Yu J, Ohuchida K, Mizumoto K, Sato N, Kayashima T, Fujita H, Nakata K, Tanaka M: MicroRNA, hsa-miR-200c, is an independent prognostic factor in pancreatic cancer and its upregulation inhibits pancreatic cancer invasion but increases cell proliferation. Mol Cancer. 2010, 9: 169-10.1186/1476-4598-9-169.
Laios A, O'Toole S, Flavin R, Martin C, Kelly L, Ring M, Finn SP, Barrett C, Loda M, Gleeson N, D'Arcy T, McGuinness E, Sheils O, Sheppard B, O' Leary J: Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Mol Cancer. 2008, 7: 35-10.1186/1476-4598-7-35.
Hu X, Macdonald DM, Huettner PC, Feng Z, El Naqa IM, Schwarz JK, Mutch DG, Grigsby PW, Powell SN, Wang X: A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecol Oncol. 2009, 114: 457-464. 10.1016/j.ygyno.2009.05.022.
Eitan R, Kushnir M, Lithwick-Yanai G, David MB, Hoshen M, Glezerman M, Hod M, Sabah G, Rosenwald S, Levavi H: Tumor microRNA expression patterns associated with resistance to platinum based chemotherapy and survival in ovarian cancer patients. Gynecol Oncol. 2009, 114: 253-259. 10.1016/j.ygyno.2009.04.024.
Flavin R, Smyth P, Barrett C, Russell S, Wen H, Wei J, Laios A, O'Toole S, Ring M, Denning K, Li J, Aherne S, Sammarae D, Aziz NA, Alhadi A, Finn SP, Loda M, B S, Sheils O, O'Leary JJ: miR-29b expression is associated with disease-free survival in patients with ovarian serous carcinoma. Int J Gynecol Cancer. 2009, 19: 641-647. 10.1111/IGC.0b013e3181a48cf9.
Shi XB, Xue L, Yang J, Ma AH, Zhao J, Xu M, Tepper CG, Evans CP, Kung HJ, deVere White RW: An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc Natl Acad Sci USA. 2007, 104: 19983-19988. 10.1073/pnas.0706641104.
Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D'Urso L, Pagliuca A, Biffoni M, Labbaye C, Bartucci M, Muto G, Peschle C, De Maria R: The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008, 14: 1271-1277. 10.1038/nm.1880.
Lin SL, Chiang A, Chang D, Ying SY: Loss of mir-146a function in hormone-refractory prostate cancer. RNA. 2008, 14: 417-424. 10.1261/rna.874808.
Saini S, Majid S, Yamamura S, Tabatabai ZL, Suh SO, Shahryari V, Chen Y, Deng G, Tanaka Y, Dahiya R: Regulatory role of miR-203 in prostate cancer progression and metastasis. Clin Cancer Res. 2011, 17: 5287-5298. 10.1158/1078-0432.CCR-10-2619.
Hagman Z, Larne O, Edsjo A, Bjartell A, Ehrnstrom RA, Ulmert D, Lilja H, Ceder Y: miR-34c is downregulated in prostate cancer and exerts tumor suppressive functions. Int J Cancer. 2010, 127: 2768-2776. 10.1002/ijc.25269.
Spahn M, Kneitz S, Scholz CJ, Stenger N, Rudiger T, Strobel P, Riedmiller H, Kneitz B: Expression of microRNA-221 is progressively reduced in aggressive prostate cancer and metastasis and predicts clinical recurrence. Int J Cancer. 2010, 127: 394-403.
Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI: Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res. 2008, 14: 2588-2592. 10.1158/1078-0432.CCR-07-0666.
Lin SC, Liu CJ, Lin JA, Chiang WF, Hung PS, Chang KW: miR-24 up-regulation in oral carcinoma: positive association from clinical and in vitro analysis. Oral Oncol. 2010, 46: 204-208. 10.1016/j.oraloncology.2009.12.005.
Liu CJ, Kao SY, Tu HF, Tsai MM, Chang KW, Lin SC: Increase of microRNA miR-31 level in plasma could be a potential marker of oral cancer. Oral Dis. 2010, 16: 360-364. 10.1111/j.1601-0825.2009.01646.x.
Yang CC, Hung PS, Wang PW, Liu CJ, Chu TH, Cheng HW, Lin SC: miR-181 as a putative biomarker for lymph-node metastasis of oral squamous cell carcinoma. J Oral Pathol Med. 2011, 40: 397-404. 10.1111/j.1600-0714.2010.01003.x.
Zhao H, Shen J, Medico L, Wang D, Ambrosone CB, Liu S: A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer. PLoS One. 2010, 5: e13735-10.1371/journal.pone.0013735.
Roth C, Rack B, Muller V, Janni W, Pantel K, Schwarzenbach H: Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010, 12: R90-10.1186/bcr2766.
Wang F, Zheng Z, Guo J, Ding X: Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor. Gynecol Oncol. 2010, 119: 586-593. 10.1016/j.ygyno.2010.07.021.
Heneghan HM, Miller N, Kelly R, Newell J, Kerin MJ: Systemic miRNA-195 differentiates breast cancer from other malignancies and is a potential biomarker for detecting noninvasive and early stage disease. Oncologist. 2010, 15: 673-682. 10.1634/theoncologist.2010-0103.
Asaga S, Kuo C, Nguyen T, Terpenning M, Giuliano AE, Hoon DS: Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin Chem. 2011, 57: 84-91. 10.1373/clinchem.2010.151845.
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J, Zhang CY: Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18: 997-1006. 10.1038/cr.2008.282.
Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, Chen Y, Xu L, Zen K, Zhang C, Shen H: Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010, 28: 1721-1726. 10.1200/JCO.2009.24.9342.
Roth C, Kasimir-Bauer S, Pantel K, Schwarzenbach H: Screening for circulating nucleic acids and caspase activity in the peripheral blood as potential diagnostic tools in lung cancer. Mol Oncol. 2011, 5: 281-291. 10.1016/j.molonc.2011.02.002.
Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ: Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009, 58: 1375-1381. 10.1136/gut.2008.167817.
Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X: Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010, 127: 118-126. 10.1002/ijc.25007.
Pu XX, Huang GL, Guo HQ, Guo CC, Li H, Ye S, Ling S, Jiang L, Tian Y, Lin TY: Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol. 2010, 25: 1674-1680. 10.1111/j.1440-1746.2010.06417.x.
Zhang C, Wang C, Chen X, Yang C, Li K, Wang J, Dai J, Hu Z, Zhou X, Chen L, Zhang Y, Li Y, Qiu H, Xing J, Liang Z, Ren B, Yang C, Zen K, Zhang CY: Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem. 2010, 56: 1871-1879. 10.1373/clinchem.2010.147553.
Liu R, Zhang C, Hu Z, Li G, Wang C, Yang C, Huang D, Chen X, Zhang H, Zhuang R, Deng T, Liu H, Yin J, Wang S, Zen K, Ba Y, Zhang CY: A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer. 2011, 47: 784-791. 10.1016/j.ejca.2010.10.025.
Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, Konishi H, Morimura R, Deguchi K, Fujiwara H, Okamoto K, Otsuji E: Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. 2010, 102: 1174-1179. 10.1038/sj.bjc.6605608.
Yamamoto Y, Kosaka N, Tanaka M, Koizumi F, Kanai Y, Mizutani T, Murakami Y, Kuroda M, Miyajima A, Kato T, Ochiya T: MicroRNA-500 as a potential diagnostic marker for hepatocellular carcinoma. Biomarkers. 2009, 14: 529-538. 10.3109/13547500903150771.
Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, Zhang JF, Shen HB, Zhang CY, Zen K: Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010, 70: 9798-9807. 10.1158/0008-5472.CAN-10-1001.
Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, Huang L, Li H, Tan W, Wang C, Lin D: Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011, 50: 136-142. 10.1002/mc.20712.
Morita K, Taketomi A, Shirabe K, Umeda K, Kayashima H, Ninomiya M, Uchiyama H, Soejima Y, Maehara Y: Clinical significance and potential of hepatic microRNA-122 expression in hepatitis C. Liver Int. 2011, 31: 474-484. 10.1111/j.1478-3231.2010.02433.x.
Gui J, Tian Y, Wen X, Zhang W, Zhang P, Gao J, Run W, Tian L, Jia X, Gao Y: Serum microRNA characterization identifies miR-885-5p as a potential marker for detecting liver pathologies. Clin Sci (Lond). 2011, 120: 183-193. 10.1042/CS20100297.
Qu KZ, Zhang K, Li H, Afdhal NH, Albitar M: Circulating MicroRNAs as Biomarkers for Hepatocellular Carcinoma. J Clin Gastroenterol. 2011, 45: 355-360. 10.1097/MCG.0b013e3181f18ac2.
Hausler SF, Keller A, Chandran PA, Ziegler K, Zipp K, Heuer S, Krockenberger M, Engel JB, Honig A, Scheffler M, Dietl J, Wischhusen J: Whole blood-derived miRNA profiles as potential new tools for ovarian cancer screening. Br J Cancer. 2010, 103: 693-700. 10.1038/sj.bjc.6605833.
Lodes MJ, Caraballo M, Suciu D, Munro S, Kumar A, Anderson B: Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS One. 2009, 4: e6229-10.1371/journal.pone.0006229.
Moltzahn F, Olshen AB, Baehner L, Peek A, Fong L, Stoppler H, Simko J, Hilton JF, Carroll P, Blelloch R: Microfluidic-based multiplex qRT-PCR identifies diagnostic and prognostic microRNA signatures in the sera of prostate cancer patients. Cancer Res. 2011, 71: 550-560. 10.1158/0008-5472.CAN-10-1229.
Brase JC, Johannes M, Schlomm T, Falth M, Haese A, Steuber T, Beissbarth T, Kuner R, Sultmann H: Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer. 2011, 128: 608-616. 10.1002/ijc.25376.
Zhang HL, Yang LF, Zhu Y, Yao XD, Zhang SL, Dai B, Zhu YP, Shen YJ, Shi GH, Ye DW: Serum miRNA-21: elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate. 2011, 71: 326-331. 10.1002/pros.21246.
Yaman Agaoglu F, Kovancilar M, Dizdar Y, Darendeliler E, Holdenrieder S, Dalay N, Gezer U: Investigation of miR-21, miR-141, and miR-221 in blood circulation of patients with prostate cancer. Tumour Biol. 2011, 32: 583-588. 10.1007/s13277-011-0154-9.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2407/11/500/prepub
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
FF Liu is a Section Editor for BMC Cancer.
Authors' contributions
AH performed the literature review and prepared the manuscript. CH performed the literature review and participated in manuscript preparation. EI prepared the figure. FL designed and edited the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Hui, A., How, C., Ito, E. et al. Micro-RNAs as diagnostic or prognostic markers in human epithelial malignancies. BMC Cancer 11, 500 (2011). https://doi.org/10.1186/1471-2407-11-500
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2407-11-500