Abstract
Background
MeCP2, methyl-CpG-binding protein 2, binds to methylated cytosines at CpG dinucleotides, as well as to unmethylated DNA, and affects chromatin condensation. MECP2 mutations in females lead to Rett syndrome, a neurological disorder characterized by developmental stagnation and regression, loss of purposeful hand movements and speech, stereotypic hand movements, deceleration of brain growth, autonomic dysfunction and seizures. Most mutations occur de novo during spermatogenesis. Located at Xq28, MECP2 is subject to X inactivation, and affected females are mosaic. Rare hemizygous males suffer from a severe congenital encephalopathy.
Methods
To identify the pathways mis-regulated by MeCP2 deficiency, microarray-based global gene expression studies were carried out in cerebellum of Mecp2 mutant mice. We compared transcript levels in mutant/wildtype male sibs of two different MeCP2-deficient mouse models at 2, 4 and 8 weeks of age. Increased transcript levels were evaluated by real-time quantitative RT-PCR. Chromatin immunoprecipitation assays were used to document in vivo MeCP2 binding to promoter regions of candidate target genes.
Results
Of several hundred genes with altered expression levels in the mutants, twice as many were increased than decreased, and only 27 were differentially expressed at more than one time point. The number of misregulated genes was 30% lower in mice with the exon 3 deletion (Mecp2 tm1.1Jae) than in mice with the larger deletion (Mecp2 tm1.1Bird). Between the mutants, few genes overlapped at each time point. Real-time quantitative RT-PCR assays validated increased transcript levels for four genes: Irak1, interleukin-1 receptor-associated kinase 1; Fxyd1, phospholemman, associated with Na, K-ATPase;Reln, encoding an extracellular signaling molecule essential for neuronal lamination and synaptic plasticity; and Gtl2/Meg3, an imprinted maternally expressed non-translated RNA that serves as a host gene for C/D box snoRNAs and microRNAs. Chromatin immunoprecipitation assays documented in vivo MeCP2 binding to promoter regions of Fxyd1, Reln, and Gtl2.
Conclusion
Transcriptional profiling of cerebellum failed to detect significant global changes in Mecp2-mutant mice. Increased transcript levels of Irak1, Fxyd1, Reln, and Gtl2 may contribute to the neuronal dysfunction in MeCP2-deficient mice and individuals with Rett syndrome. Our data provide testable hypotheses for future studies of the regulatory or signaling pathways that these genes act on.
Similar content being viewed by others
Background
DNA methylation is one of several strategies cells employ to regulate gene expression. In mammals, cytosine methylation at CpG dinucleotides is essential for tissue-specific gene expression, X-chromosome inactivation, genomic imprinting and silencing of transposable elements. The methylation signal is interpreted by a family of proteins that bind to methylated CpGs and recruit transcriptional repressor complexes. MeCP2, methyl-CpG-binding protein 2, was the first identified member of this family [1]. MeCP2 is an abundant nuclear protein that binds preferentially to heterochromatin regions on mouse chromosomes [1]. MeCP2 has a methyl-CpG-binding domain (MBD) that binds to methylated CpG dinucleotides adjacent to AT sequences [2]. Chromatin immunoprecipitation (ChIP) with anti-MeCP2 antibodies precipitated a large number of repetitive sequences as well as gene sequences [3, 4]. In vitro, MeCP2 binds to promoters and represses transcription of diverse disease-related loci, including FMR1, H19, CCNA1, BRCA1, and several other genes [5–8]. Additional functions of MeCP2 may be unrelated to mCpG binding: chromatin compaction, chromatin looping, alternative splicing, and nuclear reorganization [4, 9–12].
MECP2 mutations are identified in ~ 90% of cases of classical Rett syndrome (RTT) [13–15]. RTT (OMIM 312750) is a neurological disorder characterized by apparently normal development for the first few months followed by developmental stagnation and regression, loss of purposeful hand movements and speech, truncal ataxia, stereotypic hand movements, deceleration of brain growth, autonomic dysfunction and seizures [16–19]. MECP2 is located at Xq28 and is subject to X inactivation. The classical RTT phenotype is manifest in heterozygous females that are mosaics for cells with either normal or mutant/absent MeCP2 protein. Males with a deleterious MECP2 mutation have no functional MeCP2 protein. Although apparently normal at birth, they display stagnant postnatal development, irregular respiration, central hypoxia and early death [20]. Currently, there is no effective therapy to prevent the developmental regression, autonomous nervous system dysfunction and loss of communication skills in RTT individuals. Gaining more information about MeCP2's functions and identifying the regulatory circuits that are disrupted by MeCP2 deficiency are essential steps for understanding the pathophysiology of RTT.
Originally, it was hypothesized that MeCP2 is a master regulatory protein that prevents inappropriate expression of a potentially large set of genes, and that in the absence of MeCP2, the expression of many genes would be altered. This hypothesis was not confirmed by microarray-based global expression studies. The numbers of mis-regulated genes were quite limited in different tissues from RTT individuals and mouse models with MeCP2 loss-of-function mutations. The lists of mis-regulated genes from different studies are essentially non-overlapping, and the search for specific MeCP2-target genes has turned up very few validated candidates [21–27].
We aimed to extend previous global gene expression studies in Mecp2 mutant mouse models by focusing on the cerebellum that was excluded in the previous work of Tudor et al. [23]. Gene expression patterns in the cerebellum are quite distinct from those in other brain regions in many mouse strains studied by Nadler et al. [28]. Expression of Mecp2 increases postnatally and is abundant in the adult cerebellum [29]. The truncal tremor observed in Mecp2-deficient mice [30, 31] and the ataxia observed in RTT girls suggest cerebellar pathology. In mutant Mecp2-/Y mice, the cell bodies of cerebellar granule neurons are smaller and more densely packed [30]. Postmortem examination of the cerebella of five RTT females ranging in age from 7–30 years revealed loss of Purkinje cells, atrophy and gliosis [32]. These published data provided the rationale for our study design. To address the possibility of mutation-specific and developmental time-specific changes, we compared expression changes in males of two different Mecp2-mutant mouse strains and at three different postnatal time points.
Consistent with previous studies, we did not detect substantial up-regulation of a discrete set of genes. However, we observed moderately increased transcript levels mostly in 8 wk old mutant mice. For four genes, Irak1, Fxyd1, Reln, and Gtl2, increased transcript levels were validated by real-time quantitative RT-PCR. Attempts at replication of the data on independent samples and in different brain regions yielded variable results. Physical association of these genes with MeCP2 was confirmed by chromatin immunoprecipitation (ChIP) assays that documented MeCP2 binding to promoter regions of Fxyd1, Reln, and Gtl2. This study reports, for the first time, cerebellar expression profiles in Mecp2-mutants and proposes novel MeCP2 target genes that may contribute to the pathophysiology observed in RTT individuals and MeCP2-deficient mice.
Methods
Mouse breeding and sample collection
Females heterozygous for the J-allele (Mecp2 tm1.1Jae) [30] were obtained from Rudolf Jaenisch and bred to BALB/cJ males. Females heterozygous for the B-allele (Mecp2tm1.1Bird) [31] were purchased from the Jackson Laboratory and were bred to C57BL/6J male mice. Tail DNA samples were genotyped for Mecp2 and Sry (for sex determination) as described [33]. Sry primers: F: TCCCAGCATGCAAAATACAG; R: TGGTCATGAAACTGCTGCTT. Only male mutant mice and their male wild-type siblings were used, because female heterozygotes are mosaic for MeCP2 deficiency and have variable expressivity of the mutant phenotype. For the microarray experiments and technical validation by qRT-PCR, we studied 23 mutants and 25 wild-type J-allele mice, and 32 mutant and 34 wild-type B-allele mice. For biological validation by qRT-PCR of a new set of samples, we used 20 mutant and 16 wild-type B-allele mice. Animals were euthanized with CO2. Cerebella and forebrains were snap frozen on dry ice. Total RNA was extracted using TRIzol Reagent (Invitrogen) and analyzed on a 1.5% agarose gel in MOPS buffer. RNA was concentrated and purified with YM30 microcon columns (Millipore). The Animal Care Committee of Stanford University approved all experimental procedures.
Human brain samples
Brain samples from RTT individuals, including two hemizygous males with known MECP2 mutations, and non-RTT controls were obtained from the NICHD Brain and Tissue Bank for Developmental Disorders (UMB# 1238, 1420, 1748, 1815, 4516, and 4852) and from the Harvard Brain Tissue Resource Center (#b4160). We extracted total RNA from frozen frontal cortex using RNA Stat 60 (Tel-Test) or TRIzol reagent (Invitrogen). All human materials were obtained and studied under a protocol approved by the Stanford Human Research Protection Program.
Expression cDNA microarrays
Microarrays were obtained from the Stanford Functional Genomics Facility (SFGF) [34]. These arrays contain 42,000 different transcripts that represent over 26,000 different genes. The transcripts vary in size and are PCR products from the RIKEN FANTHOM clone sets and a consortium of Stanford collaborators. The clone IDs on the cDNA microarrays are available from SFGF. Further information on the clones can be found at SOURCE [35].
cDNA labelling and microarray hybridization
For preparation of reference RNA, we used a pool of 100 cerebella collected from wild-type C57BL/6J and BALB/cJ male mice ranging from P7 to P60 in age. Reference RNA (20 μg) was reverse transcribed using an anchored oligo(dT) primer and labeled with Cy3-dCTP using the CyScribe First-Strand cDNA Labeling Kit (Amersham Biosciences, Catalog # RPN6200). Total RNA (20 μg) from wild-type samples or mutant samples was reverse transcribed and labeled with Cy5-dCTP. Following purification with YM30 microcon tubes, labeled reference cDNA was combined with either labeled wild-type cDNA or labeled mutant cDNA. The combined reactions (reference and wild-type or reference and mutant sample) were applied to a cDNA microarray. Hybridizations were carried out at 65°C for 14–16 hr in a waterbath or hybridization oven. Blocking reagents were added according to the Brown Lab protocol [36].
Microarray data analysis
Following hybridization, microarrays were washed and immediately scanned with a GenePix 4000B microarray scanner (Axon Instruments/Molecular Devices). Automatic and manual quality analysis of array spots was performed with Genepix Pro 4.0. All microarray data were deposited in the Stanford Microarray Database and normalized by total intensity. The Log (base2) of Red/Green Normalized Ratio (Mean) for each gene was retrieved and averaged only if the spot on the microarray was from the same microtiter plate (LUID – laboratory microtiter well ID). Genes were centered by the mean. Up to five replicate microarrays were evaluated for each mutant allele at each time point. There are 41,000 different spots on each microarray. Some of those spots are not "good" spots, meaning they do not pass the Genepix program filter and/or our manual evaluation. Spots do not pass our filter if the signal is too low, the spot does not have an even distribution of signal, or if there are any other experimental problems that deem the spot of poor quality. Some spots removed for low signal may imply that the gene is not expressed. Regardless of the reason, genes were excluded from our analysis if more than 85% of the spots representing a particular gene are filtered from our data set across replicate microarrays. After filtering, the number of genes that remained in our data set were: J-allele: 2 wk (25,365), 4 wk (30,657), 8 wk (29,774); B-allele: 2 wk (28,939), 4 wk (28,005), 8 wk (26,473). Differentially expressed genes were defined as genes whose expression is consistently (P < 0.05) altered across microarray experiments. Paired t-tests (Microsoft Excel) were performed by comparing the expression of each gene in a mutant sample to expression in its wild-type sibling. The data were analyzed with Microsoft Excel and SAM (Sequence Analysis of Microarrays) software [37]. Microarrays were centered and clustered using hierarchical clustering software available at the Stanford Microarray Database [38, 39]. The cDNA microarray data can be accessed online via the SMD [88] or via the EBI ArrayExpress [89].
Real-time quantitative RT-PCR (qRT-PCR)
Total RNA (2 μg) was treated with 2U DNase I (Ambion) and reverse transcribed with random hexamers as primers and Superscript II (Invitrogen). qRT-PCR was performed using an ABI 5700 (AME Bioscience) instrument. Melting curve analysis was performed for each reaction to ensure a single peak. Amplicons were visualized after electrophoresis on a 2% agarose gel to ensure presence of a single amplicon. Amplification of β-actin, β 2-microglobulin, Rps28 and RPS18 transcripts served as RNA controls for relative quantitation (User Bulletin #2, Applied Biosystems). Mouse primers used (5' – 3'):
Mecp2 (F: TGACTTCACGGTAACTGGGAG; R: TTTCACCTGAACACCTTCTGATG),
β-actin (F: TGACCCTGAAGTACCCCATTGA; R: CCATGTCGTCCCAGTTGGTAAC)
β 2m (F: ACCCGCCTCACATTGAAATCC; R: CGATCCCAGTAGACGGTCTTG),
Rps28 (F: TAGGGTAACCAAAGTGCTGGGCAG; R: GACATTTCGGATGATAGAGCGG),
Irak1 (F: CAGAACCACCACAGATCATCATC; R: GGCTATCCAAGACCCCTTCTTC),
Fxyd1 (F: TCCATTCACCTACGATTACCACA; R: GAATTTGCATCGACATCTCTTGC),
Reln (F: CTGTGTCATACGCCACGAACA; R: GGGGAGGTACAGGATGTGGAT),
Gtl2 (F: GACCCACCTACTGACTGATGAACTG; R: GTGAAGACACAACAGCCTTTCTCC).
Human primers used (5' – 3'):
FXYD1 (F: AGGGACAATGGCGTCTCTTG; R: CGTAAGTGAACGGGTCGTGTT (NM_005031)
RPS18 (F: CTTTGCCATCACTGCCATTA; R: ACACGTTCCACCTCATCCTC)
Chromatin Immunoprecipitation (ChIP)
We modified a protocol kindly provided by P. Farnham [40]. Brains of 4 wk-old male mice were isolated and minced to small pieces. Cross-linking reactions were performed at room temperature for 15 min in the presence of 1% formaldehyde in PBS. Nuclear extracts were prepared and chromatin was fragmented by sonication to produce DNA fragments of an average length between 500 bp and 1000 bp. After pre-cleaning with salmon sperm DNA/protein A agarose (Upstate), we incubated the nuclear extracts with anti-MeCP2 antibody (Upstate) at 4°C overnight. MACs Protein A Micro Beads (Miltenyi Biotec) were added to the reaction to allow the formation of magnetically labeled immune complexes. We purified the bound complexes using a microcolumn following the manufacturer's instructions. After reversal of cross-links and digesting with proteinase K, we isolated DNA fragments with a DNA purification kit (Qiagen). We performed 36 cycles of PCR with primers specific to the first ~ 500–1000 bp of the gene promoter region or to the DMR at the 5' region of the Gtl2 gene.
Primer pairs: Fxyd1 (F: GCATGTCCTGCTGTATGTCG; R: CAGCCAGGGTCAAGAAATGT), Reln (F: AAAGGGAGATTGGGTGACG; R: ACGTGCTTCTGGATGGTTTC), Gtl2 – A (F: TCCAACACGAAATTCTGCAA; R: TGCTGGTCAACATGAACCTC) amplifying nt 94579–94964 and the nested primers Gtl2 – B (F: CGGTCCACTAGGGCTTTTT; R: CCAGGTTTTTAGACCCCAGA) amplifying nt 94645–94775 of AJ320506.
Results
Mecp2-mutant mice differ in phenotype and expression of mutant Mecp2 allele
We examined gene expression in the cerebellum of two different Mecp2-mutant mouse models. The Mecp2 tm1.1Jae (J-allele) mutant has an in-frame deletion of exon 3 that codes for half of the MBD [30] (Figure 1A). Therefore, the expected mutant protein, if present, would be unable to bind methylated DNA. In contrast, the sequences encoding the transcriptional-repression domain (TRD), nuclear localization signals and C-terminal WW-binding domain, all encoded by exon 4, are retained. The second mutant, Mecp2 tm1.1Bird (B-allele), is deleted for exon 3 and the coding sequence of exon 4 [31]. In this allele, ~ 97% of the Mecp2-coding sequence is deleted, as well as a small part of the 3'UTR. No mutant mRNA was detected on Northern blot [31].
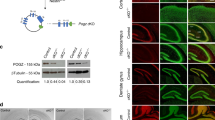
Structure and expression of Mecp2 mutant alleles. A. Genomic structure of Mecp2 mutations . a. Wild-type Mecp2 gene. MBD, methyl-CpG binding domain; TRD, transcription repression domain. White or hatched: coding sequence. Grey: non-coding parts of transcript. Exons 1 and 2 that are alternatively spliced, and intron 2, are not drawn to scale. Horizontal bar above exon 4 denotes location of the amplicon analyzed by qRT-PCR (in Figure 1B) b. The Mecp2 tm1.1Jae (J-allele) mutant has a deletion of exon 3 that encodes the N-terminal half of the MBD. c. In the Mecp2 tm1.1Bird (B-allele) mutant, exon 3, the coding portion of exon 4 and part of the 3'UTR are deleted. This larger deletion eliminates ~ 97% of the coding sequence. B. Real-time quantitative RT-PCR analysis of Mecp2 expression in cerebellum . Using primers within the deleted region, Mecp2 transcript was undetectable in mutant mice with the B-allele, when four wild-type and four mutant samples were compared at 8 wk. This proves the specificity of the primers. In mice with the J-allele, mean Mecp2 expression was higher in the mutants, but this difference was not statistically significant (P = 0.11), when six wild-type and seven mutant samples were compared at 8 wk.
Asymptomatic females, heterozygous for a Mecp2 mutation and less than 6 months old, were bred to wild-type males. Offspring were genotyped for Mecp2 and Sry (for sexing). Cerebellar RNA was extracted from hemizygous males at mean ages of 2 wk, 4 wk and 8 wk. At 4 wk, 22% of mutant B-allele mice but none of the J-allele mice were symptomatic. Differences in body weight were observed in both sets of mice beginning at 4 wk, with the mutants being significantly smaller than wild-type littermates. At the "8 wk" time point, 80% of B-allele mice were symptomatic, while the range for J-allele mice extended from 10% at 7 wk to 71% at 9 wk. The most consistent abnormality observed in mutant mice was decreased mobility. At later stages, they developed tremors and hind-limb clasping and appeared poorly groomed.
Primers designed to amplify a region of exon 4 did not detect Mecp2 transcripts in B-allele mice as expected, but qRT-PCR studies revealed normal levels of the mutant Mecp2 transcript in 8 wk old mice with the J-allele (Figure 1B). Therefore, while the Mecp2 tm1.1Bird B-allele represents a true null mutation, the Mecp2 tm1.1Jae (J-allele) produces a stable mutant mRNA capable of directing synthesis of an internally deleted mutant protein that may have retained functions unrelated to mCpG binding. By using whole brain extracts for Western blot analyses, we were unable to detect the predicted protein product of 309 amino acids made from the J-allele. All antibodies we tested cross-reacted with an ~ 55 kD unknown protein in all samples and, therefore, the putative J-allele product may have been obscured. Alternatively, it may have been degraded in the majority of neurons.
Differentially expressed genes in Mecp2-mutant mice
To discover gene expression differences between mutant and wild-type cerebella, we performed microarray experiments of both mutant strains under the same conditions. Our microarrays contained 26,000 different cDNA clones spotted on glass slides (Stanford Functional Genomics Facility). Reverse-transcribed cDNA samples from either wild-type or mutant cerebella were hybridized to a cDNA microarray together with a reference sample consisting of pooled cerebellum cDNA. By carrying out hierarchical clustering of the microarray data, we found that several parameters greatly influenced the clustering. In comparing expression data from B-allele mice at 8 wk of age, littermates tended to cluster more than mutants of different litters (Figure 2). Clustering data from only the wild-type control mice illustrate the effects of age, strain background, and array batch (Figure 3). Therefore, in the final analysis, we only compared mutant expression arrays to wild-type arrays of mice that were from the same litter and only microarrays from the same batch. Pair-wise t-tests were performed to combine data from different wild-type/mutant pairs.
Hierarchical clustering of microarray data from six mutant/wildtype sib pairs. Male B-allele mutants at 8 wk of age, when most of them are symptomatic, tended to cluster with their wt sibs rather than with mutants of other litters. Litters are identified by A – F. The array of the litter B wild-type sib did not pass quality control standards.
Hierarchical cluster analysis of microarray data from wild-type sibs at three different ages. Strain background had the greatest influence on clustering. BALB/c is the major background of the J-allele mice, while B-allele mice are on a C57BL/6 background. The effects of array batch, and to a lesser extent, age, are also apparent. We, therefore, compared only microarrays from littermates, at the same age and using the same array batch, in our search for differentially expressed genes.
Consistent with previous expression microarray studies of whole brain or other brain regions, we found that loss of MeCP2 function does not lead to global mis-regulation of gene expression in cerebellum. Even without correction for multiple testing, the total number of differentially expressed genes with a p-value of < 0.05 was not significantly greater than would be expected by chance (Table 1). In B-allele mice, the number of genes with increased expression was slightly greater than would be expected by chance, but only at the 8 wk time point when more of the mutant mice used were symptomatic. The majority of changes reflect an increase of transcript levels, consistent with a relaxation of gene repression.
To identify genes of potential biological significance, we studied the datasets from different time points and different alleles for common differentially expressed genes (DEG), but very few consistent differences were detected. The degree of overlap at 2 wk, 4 wk, and 8 wk for each mutant Mecp2 allele is very small (Figure 4A, Additional file 1). In male mice with the J-allele, no genes showed altered expression at all three time points. In B-allele mice, only one Riken clone (H3142G05) [35] was differentially expressed at all time points. Expression of this gene, later identified as Tep1, was increased on microarrays: 1.24, 1.52, and 1.25 fold at 2 wk, 4 wk and 8 wk, respectively. Tep1, which codes for telomerase-associated protein 1, was first discovered in Tetrahymena and is conserved from ciliates to humans. TEP1 is the RNA-binding protein of the telomerase ribonucleoprotein complex that also contains the catalytic reverse transcriptase subunit and an RNA component that serves as a template for the synthesis of telomeric repeats [41]. More recently, TEP1 was also identified as a protein component of vaults that are ribonucleoprotein complexes localized in the cytoplasm of cholinergic nerve terminals close to synaptic vesicles [42]. Specifically, TEP1 is required for the stable association of the vault RNA with the vault complex [43]. Tep1 expression was not significantly changed in mutant mice with the J-allele.
Differentially expressed genes (DEG) in cerebellum microarray study. A. Overlap of DEG at 2 wk, 4 wk, and 8 wk time points. For both mutant alleles, genes whose transcripts were either increased (1.2-fold or higher) or decreased (0.8-fold or lower) at P < 0.05 are included. Transcripts in the overlap regions are described in Additional file 1. B. Overlap of DEG across both mutant alleles at three time points . The same criteria as in Figure 4A were applied. C. Overlap of DEG across all ages in both mutant strains. The 39 transcripts include the 15 overlapping DEG from Figure 4B as well as genes that were significantly changed at one age in one mutant and at another time point in the other mutant. The list is provided in the Additional file 1.
A total of 27 genes were differentially expressed at two different time points, four in the J-allele mutants and 23 in the B-allele mutants (Figure 4A). The gene displaying the greatest expression change is Gtl2 (Gene-trap locus 2) that was up-regulated 1.6 fold at 2 wk and 1.7 fold at 8 wk in B-allele mutants. Increased RNA levels were detected by two different clones on the cDNA microarrays at 2 wk (2210008A22) and 8 wk (1110014G20). The sequences are non-overlapping, but both represent the Gtl2 locus. Gtl2 is an imprinted gene producing a non-coding RNA of unknown function. The paternally expressed gene Delta-like 1 (Dlk1) and the maternally expressed Gtl2 gene are close to each other within an imprinted gene cluster on distal mouse chromosome 12 [44]. Dlk1 expression was not significantly changed in either mutant.
Since any expression changes that are relevant for the phenotype might be seen in both mutant strains, we determined the number of common expression changes between the two strains. In general, there is a low degree of overlap (39 out of 3285 genes) between DEGs in the B- and J-allele mutants, and only 15 genes overlap at the same time points (Figure 4B, Additional file 1). Potentially functionally relevant expression changes were observed in both mutants for several genes: Smarcb1, Satb1, Fubp3 and Abt1. Smarcb1 is a component of the SWI/SNF ATP-dependent chromatin-remodeling complex. Two studies have linked MeCP2 and the SWI/SNF chromatin-remodeling complex [5, 45], although this connection is controversial [46]. SATB1 (special AT-rich sequence binding protein 1) is predominantly expressed in thymocytes and specifically binds to nuclear matrix attachment regions (MARs) [47]. It is involved in chromatin loop formation [48] and mediates formation of a higher-order transcriptionally active chromatin structure in activated T cells [49]. Satb1 is also expressed in spinal cord and distinct regions of the developing mouse brain in a mutually exclusive fashion with its close relative Satb2 [50]. Like SATB1, MeCP2 also binds to nuclear matrix-associated DNA repeat sequences in the human genome, and the chicken ortholog of MeCP2 is the MAR-binding protein ARBP [51]. In both mutant mouse models, increased expression was observed for two transcriptional activators, Fubp3 and Abt1, at 8 wk and for two other transcriptional regulators, Rab8a and Prdm4, at 8 wk and 2 wk, respectively (Additional file 1).
Increased expression of Irak1, Fxyd1, Reln, and Gtl2 in Mecp2-mutant mice
For further studies, we selected four genes based on a fold change of at least 1.4 and consistency in the directional change over the majority of microarrays: Irak1, Fxyd1, Reln, and Gtl2 (Table 2). Expression of Irak1, encoding interleukin-1 receptor-associated kinase 1, was increased in all mutant/wild-type comparisons at 8 wk in B-allele mice. In real-time qRT-PCR experiments, Irak1 expression was increased 3.5-fold in the mutant relative to the wild-type 8 wk B-allele cerebellum samples and 1.9-fold in the forebrain (RC and RF, Figure 5). In the microarray data, Irak1 expression was not significantly altered at other ages, nor was it altered in J-allele mice. The Irak1 gene is located only 3 kb distal to Mecp2. Therefore, this expression change may reflect the altered genomic environment. For example, an element that negatively influences Irak1 expression could be deleted in the B-allele, but not in the J-allele that has a much smaller deletion.
Microarray and qRT-PCR results for select DEGs. Irak1, Fxyd1, Reln and Gtl2 were selected for validation of expression changes by qRT-PCR analyses in B-allele mice. Cerebellum microarray expression data (M) are compared with qRT-PCR data from cerebellum (RC), forebrain (RF) and hippocampus (RH). For RC of Irak1, Fxyd1 and Gtl2, the 8 wk samples from the microarray analysis were used. RC2 data are derived from an independent set of litters at 7.5 to 8 wk. Error bars represent SEM. One asterisk indicates P < 0.05, two asterisks indicate P < 0.001. Irak1 expression was markedly increased in the cerebellum of 8 wk B-allele mice on microarray (2-fold, P = 0.011, four mutant/wild-type pairs) and qRT-PCR (3.5 fold, P < 0.001, four mutant/wild-type pairs) as well as in the forebrain of a different set of mice at 7.5 wk (1.9 fold, P < 0.05, three mutant/five wild-type). Fxyd1 expression was elevated 1.4-fold on microarrays (P < 0.05, five mutant/wild-type sibs) and qRT-PCR (RC) ( P = 0.053, four mutant/wild-type sibs). The RC2 data (five mutant/three wild-type) were more variable and yielded P = 0.16. Reln displayed an ~ 1.5 fold increase in expression on microarrays (P = 0.03, four mutant/wild-type pairs) and qRT-PCR (P = 0.06, five mutant/wild-type pairs) at 2 wk (RC) and was not changed in hippocampus (RH). On microarray, Gtl2 expression was increased at 8 wk, but by qRT-PCR we found highly variable Gtl2 expression in cerebellum (RC and RC2). Gtl2 expression was not significantly increased (P = 0.29, eight mutant/eight wild-type) in the forebrain.
Fxyd1 encodes the FXYD domain containing ion transport regulator 1, also called phospholemman. Fxyd1 expression was consistently increased in all microarray experiments (average 1.4 fold) of B-allele mice at 8 wk (Table 2). The results were confirmed by qRT-PCR analysis on the same cerebellum samples (RC, Figure 5). Biological replication by qRT-PCR on a different set of B-allele mice (RC2; 3 wt, 5 mut) yielded more variable results between litters resulting in large SEM error bars and a p-value of 0.13. Fxyd1 expression in forebain samples from six different litters showed no difference between mutant and wild-type samples, but Fxyd1 expression is normally very low in forebrain.
Reln, encoding the extracellular signaling molecule reelin, is highly expressed in the developing cerebellum. Reelin is essential for proper neuronal lamination and synaptic plasticity. We found increased Reln transcript levels (1.5 fold) in four microarray experiments of B-allele samples at 2 wk (M, Figure 5). Reln expression was also increased in qRT-PCR experiments (RC), but this increase is not statistically significant as there was substantial variation among the samples tested (Figure 3). No difference was seen in hippocampus when 5 wild-type and 7 mutant samples were compared (RH, Figure 5).
Gtl2, encoding an imprinted maternally expressed non-translated RNA, is the mouse homolog of MEG3, maternally expressed gene 3, in humans. A study evaluating the transcripts encoded by Gtl2 identified 13 different splice variants [52]. Our microarray data revealed significantly increased Gtl2 expression at 2 wk and 8 wk in B-allele mice (M, Figure 5). By qRT-PCR assays, the expression increase was present, but not significant in the same samples (RC) and in forebrain (RF, Figure 5). Cerebellum studies of a new set of mice (3 wt, 5 mut), however, showed no difference (RC2, Figure 5).
MeCP2 binds to the promoters of Fxyd1 and Reln and the promoter and differentially-methylated region of Gtl2 in vivo
To determine if MeCP2 binds directly to the potential target genes, we performed chromatin immunoprecipitation (ChIP) assays on brain tissues. ChIP involves cross-linking genomic DNA to proteins that are bound to it. Subsequently, DNA-protein complexes are isolated by immunoprecipitation with a MeCP2 antibody and cross-links are reversed. The MeCP2 bound sequences are used as templates for semi-quantitative PCR to determine whether the gene region of interest is binding to MeCP2 [40].
When we used primers specific for the 500–1000 bp Fxyd1 or Reln promoter regions, ChIP results indicated specific MeCP2 interaction in normal brain extracts (Figure 6). Amplification was enriched only in the presence of MeCP2 antibody (Figure 6, lanes 4). There was no, or much less, amplification of the DNA fragments when ChIP was performed either with nuclear extract from Mecp2-mutant mouse brain (lanes 1), or without MeCP2 specific antibody (lanes 2), or with non-specific antibody (IgG) (lanes 3). The results suggest that MeCP2 is part of a complex that is located at the promoter regions of these genes in normal brain and that this complex may not form in brain of Mecp2 tm1.1Bird mutant mice.
Chromatin immunoprecipitation (ChIP) analysis of differentially expressed genes. Whole brain nuclear extracts from 4 wk old wild-type and B-allele mutants were used for ChIP. Lane M, 100 bp ladder; lane 1, Mecp2-mutant chromatin precipitated with anti-MeCP2 (negative control); lane 2, wild-type chromatin precipitated with no antibody present; lane 3, wild-type chromatin precipitated with rabbit IgG, lane 4: wild-type chromatin precipitated with anti-MeCP2, lane 5: input DNA, lane 6: no DNA control. The precipitated DNA was amplified with primers specific for the promoter regions of Fxyd1 and Reln, and for the promoter/DMR of Gtl2. The Gtl2-B amplicon is located within the Gtl2-A amplicon. Precipitation with anti-MeCP2 shows enrichment relative to the IgG and no antibody controls, indicating that MeCP2 is associated with the designated regions in vivo. As negative controls, PCR was performed using primers covering regions that do not interact with MeCP2 for each ChIP reaction (not shown).
Gtl2 is an imprinted transcript whose maternal-specific expression is controlled by a differentially-methylated region (DMR) that overlaps the promoter region. MeCP2 binding was systematically analyzed using overlapping primer sets, each of which covers 250–400 bp of a region that contains the Gtl2-DMR (AJ320506, nt 92340–95350). MeCP2 binding activity was detected upstream of exon 1 and extending through exon 1 into intron 1 (AJ320506, 93720–95350) (Figure 6). This 1.6 kb region contains 63 CpG dinucleotides, many of which are differentially methylated.
FXYD1expression in human brain with MeCP2 deficiency
Recently, Deng et al (2007) [53] identified FXYD1 as a target of MeCP2 by increased expression in the frontal cortex of RTT individuals and of Mecp2-mutant mice, with no changes observed in the cerebellum. While these authors confirmed by ChIP that MeCP2 binds to the FXYD1 promoter, their expression results were the opposite of our findings in mouse brain. Therefore, we examined FXYD1 expression in human brain samples from five RTT females, two males with congenital encephalopathy (CE) and documented MECP2 mutations, and seven non-RTT controls (Figure 7). We used real-time quantitative RT-PCR with the same primers as Deng et al. [53]. We normalized FXYD1 expression levels to RPS18 expression rather than to rRNA expression. The ages of the donors differed only slightly, with controls in their 30 s and RTT females in their 20 s. The two CE males were between 1 and 2 years old. No MECP2 mutation had been identified in the single female RTT sample (UMB# 1748) with an increased FXYD1 transcript, but a brain contusion lesion was seen on autopsy. Our results do not confirm an increase in FXYD1 expression in frontal cortex of MeCP2-deficient humans, and they agree with our data on Mecp2-mutant mice. If MeCP2 were responsible for silencing FXYD1 in the forebrain, one would expect to observe a major increase in hemizygous males with CE who have no MeCP2 activity at all, as compared to RTT females who are mosaic for MeCP2-deficient and normal cells.
Relative FXYD1 expression in human frontal cortex. Quantitative real-time RT-PCR analyses were carried out on frozen frontal cortex samples from seven unaffected controls (C1 – C7), two MECP2-mutant males with congenital encephalopathy (CE) and five females diagnosed with RTT, with or without identified MECP2 mutations. The mutant samples are identified by their brain bank numbers. FXYD1 expression was normalized to RPS18, a constitutive ribosomal protein gene that is expressed at a similar level in brain. To calculate relative fold changes, control sample C4 was arbitrarily set at 1. All samples were run in triplicate on the same plate.
Discussion
The aim to discover abnormally expressed genes in MeCP2-deficient mice is two-fold: to reveal direct targets of MeCP2 modulation and to identify components of neuronal pathways and circuits that are deregulated in affected brain regions. The ultimate goals are to understand the pathophysiologic processes that lead to morbidity in RTT and to design rational therapies to prevent or revert the functional abnormalities. To reduce microarray data noise caused by cellular heterogeneity of tissues, we focused on the cerebellum, a part of the brain not previously studied. We compared expression profiles in two different MeCP2-deficient mouse strains that differ on the molecular level, the Mecp2 tm1.1Bird (B-allele) and Mecp2 tm1.1Jae (J-allele). Furthermore, we compared expression profiles at three postnatal ages that span different developmental stages in the cerebellum. At two weeks, granule cells are still mitotic and migrating to their final position in the internal granule layer. At four and eight weeks, granule cells are post-mitotic and have begun forming synapses with neurons in the cerebellum and other brain regions.
Consistent with previous studies on forebrain and hippocampus samples by Tudor and colleagues [23], we did not observe major expression changes of a large number of genes in the cerebellum. MeCP2-deficiency does not significantly alter the cerebellar transcriptome on a global scale. This is remarkable given that the vast majority of cells in the cerebellum are granule cells that had shown morphological abnormalities in J-allele mice [30]. Any significant overexpression of genes specifically in granule cells should have been detectable, as it would not be averaged out by cell type heterogeneity. Our results should caution investigators who engage in cell-type specific profiling studies. Also, no large (> 3 fold) changes were detected for any single gene. The changes in cerebellum transcript levels tended to be small (1.5 – 2 fold), but increases greatly outnumbered decreases. Overall, a number of genes with increased transcript levels greater than expected by chance were observed only in B-allele mice at 8 wk. There are several possible explanations for these findings.
First, the mutations are different. While B-allele mice are null at the Mecp2 locus, the J-allele mutation consists of an in-frame deletion of exon 3 that produces a stable mutant transcript. The putative mutant protein, which lacks the MBD domain, but has an intact nuclear localization signal, transcription repression domain and C-terminal region, may have maintained certain functions. Some evidence has been reported for a truncated MeCP2 protein in the J allele mice. Luikenhuis et al. [54], saw diffuse nuclear staining with a MeCP2 antibody and speculated that this probably arose from the detection of the truncated protein. This interpretation is in line with the original report of the J-allele mice [30], where Western blot showed a smaller band that was interpreted to represent the truncated protein, even though the wild-type samples had a protein of apparently the same size, but at a lower level of intensity. The possibility cannot be excluded, however, that the diffuse nuclear staining was caused by a cross-reacting protein that does not localize to DAPI-positive foci. Using tissue microarrays to study the mosaic composition of Mecp2-expressing and non-expressing cells in female mice, Braunschweig et al. [55], reported a background immunoreactivity with a C-terminal antibody in male J-allele mice that was apparently not present in mice with the B-allele. These results would argue in favor of the presence of an internally deleted mutant protein present in the J-allele brain, although the intensity was rather low.
Second, the mice were on different strain backgrounds: B-allele mice on C57BL/6 and J-allele mice on a mixed background, mostly BALB/cJ. Our hierarchical clustering data clearly demonstrated a strong effect of strain background (Figure 3). Random mixed strain background noise should be overcome, to some extent, by our comparing only wild-type and mutant sib-pairs and by including a sufficiently large number of sib-pairs (16 for the B-allele and 13 for the J allele).
Third, 80% of the B-allele mice at 8 wk were symptomatic, while fewer of the J-allele mice were showing symptoms. When the disease process in the brain is further advanced, the expression changes may be caused by pathologic events far downstream of direct MeCP2 action.
Although the global level of gene expression is not significantly altered, that does not preclude the possibility that some of the genes may have changed expression levels that are biologically relevant. Therefore, we focused on whether differentially expressed genes showed a similar change in other brain regions and in a different set of mice (biological validation), which is a more relevant evaluation than "statistical" validation. We chose four genes, Irak1, Fxyd1, Reln, and Gtl2, whose expression levels were increased in the most consistent fashion. By ChIP analyses, we show that MeCP2 binds to the promoters of Fxyd1 and Reln and to a differentially methylated region of the imprinted gene Gtl2 in normal mouse brain, suggesting that these genes may be direct targets of MeCP2.
Irak1
The interleukin-1 receptor-associated kinase 1 gene revealed the largest significant expression increase of any gene, in cerebellum (2.0-fold on microarray and 3.5-fold on qRT-PCR assays) and in forebrain (1.9-fold), but only in B-allele mice. Irak1 is located ~ 3 kb downstream of Mecp2 in the same transcriptional orientation (UCSC Genome browser, Feb 2006 assembly). This suggests that a negative regulator of Irak1 could have been lost with the B-allele deletion, or that the altered local chromatin conformation leads to increased Irak1 expression. The less likely possibility that MeCP2 directly regulates the expression of Irak1 would assume that the J-allele has maintained a negative regulatory function, since Irak1 expression is not increased in J-allele mutants. This allele-specific difference raises the concern that the increased Irak1 expression may be responsible for some dysregulated genes in the B-allele mice that are not dysregulated in the J-allele mice. To identify bona fide MeCP2 targets we, therefore, focused on genes whose promoters bind MeCP2 in ChIP assays. Nevertheless, any studies done with the B-allele mice, at any level, from behavior to histology, should be considered in the light of possible downstream effects of Irak1 overexpression.
IRAK1 belongs to the ser/thr protein kinase family, pelle subfamily. The protein is comprised of a death domain in the N-terminus, a central serine/threonine kinase domain, and a C-terminal serine/threonine rich region. Once phosphorylated, IRAK1 recruits the adapter protein PELI1 and causes activation of Toll-like-receptor-mediated intracellular signaling pathways. Three different splice variants in humans and mice differ in their relative abundance in brain, and differential splicing of IRAK1 may correlate with the aging process [56]. Our qRT-PCR assay amplified all three splice forms. Given the postulated role of MeCP2 in alternative splicing [11], it might be of interest to evaluate the relative abundance of the different Irak1 splice forms in Mecp2-mutant B-allele brain.
Fxyd1
In wild-type mice, Fxyd1 expression increases drastically from 2 wk to 8 wk in the cerebellum (our microarray data). In Mecp2 mutant mice, microarray and qRT-PCR studies revealed a significant increase of Fxyd1 transcripts at the 8 wk time point in B-allele mutants when compared to wild-type littermates. ChIP analyses confirmed the binding of MeCP2 to the Fxyd1 promoter. We, thus, have identified Fxyd1 as a validated target of MeCP2 binding. In contrast to a recent report by Deng et al. [53], however, Fxyd1 transcripts were not increased in the mutant forebrain where expression levels are normally very low. Attempting to replicate the reported studies in humans, we have used qRT-PCR to measure FYXD1 expression in frontal cortex from male and female individuals with MECP2 mutations and unaffected controls. We found no evidence for increased expression in the mutant samples. Therefore, our data in humans and mice do not support the hypothesis that MeCP2 is responsible for keeping FYXD1/Fxyd1 expression low in the majority of cells of forebrain. A small increase of expression in a neuronal subtype could have gone undetected, but the large increases reported by Deng et al. [53] in RTT frontal cortex are not confirmed by our results.
The protein product, FXYD1, also called phospholemman (PLM), is a small molecule with a single transmembrane domain and a member of the FXYD family of small ion transport regulators. FXYD proteins regulate Na-K-ATPase activity in a tissue- and isoform-specific way. Each FXYD gene is expressed in one or more specific tissues [57]. FXYD1 is a major membrane phosphoprotein in heart and muscle. In brain, FXYD1 is predominantly found in the cerebellum, specifically in the molecular layer, in Purkinje neurons, and in axons traversing the granule cell layer, as well as in the choroid plexus [58]. The protein forms a helical structure and inserts into lipid membranes in a trans-bilayer fashion [59]. FXYD1 tetramers form ion channels selective for K+, Cl-, and taurine that are physically associated with the Na-K-ATPase [60]. Recent studies revealed that FXYD1 also inhibits the cardiac Na+/Ca2+ exchanger (NCX1) [61]. Overexpression of FXYD1 in adult cardiac myocytes acutely alters contractility as a function of extracellular Ca+ concentration, and Fxyd1 knock-out mice have cardiac hypertrophy [62]. Alteration of Fxyd1 expression in the heart of Mecp2-deficient mice and humans could conceivably contribute to cardiac dysfunction and sudden death.
Another member of the FXYD family, FXYD4/CHIF (corticosteroid hormone induced factor) was initially isolated as a glucocorticoid response gene [63]. Interestingly, two putative MeCP2 target genes, Sgk1 and Fkbp5 are also glucocorticoid-inducible and were found to be upregulated in 8 wk old B-allele mice studied previously [25]. Furthermore, in the Mecp2 308 mouse model of MeCP2 deficiency, circulating corticosteroid levels were increased in response to stress, and the gene for corticotropin-releasing hormone (Crh) was overexpressed in brain regions, leading to the conclusion that the mutant mice have an enhanced corticosteroid response to stress via increased CRH signaling [64]. Future studies should evaluate whether FXYD1, like FXYD4, is also corticosteroid inducible.
Reln
Our identification of Reln as a primary target of MeCP2 is supported by the following observations: Reln transcript levels were increased in microarray and qRT-PCR experiments comparing cerebella from a total of 12 wild-type and 9 mutant mice. And the expression change was seen at the earliest time point (2 wk) when all mice were asymptomatic. No expression change was found in hippocampus. ChIP assay on whole brain documented that MeCP2 binds to the Reln promoter in normal mice. The proximal promoter of Reln is highly CpG-rich and methylated in whole brain extracts [65]. Multiple hypermethylation sites at the Reln promoter can be induced by injection of L-methionine. And in a methionine-induced model of schizophrenia, MeCP2 was shown to bind to the Reln promoter [66, 67].
Reelin is a large extracellular protein that is produced by discrete populations of cells in the brain. It acts through the extracellular milieu on neighboring target cells. Until recently, the primary function of reelin was thought to involve regulation of neuronal migration during fetal brain development, by reelin providing an architectonic signal for the guidance of migrating neurons [68, 69]. Reelin-expressing cells include the Cajal-Retzius (CR) cells in the cortical marginal zone, that are the first neurons to express Mecp2 in development, and cerebellar granule cells. MeCP2-deficient mice have a thinner cortex and more densely packed neurons that could result from migratory defects.
Reln expression continues in the adult, in particular in a subset of GABAergic neurons of the forebrain, as well as in the olfactory bulb [70]. Reelin can modulate synaptic plasticity and enhance long-term potentiation (LTP) in adult hippocampal cultures [71]. The signaling pathway that reelin acts on involves two high-affinity binding receptors, the very low density lipoprotein receptor (VLDLR) and the apolipoprotein E receptor 2 (ApoER2) [72]. Binding of reelin to VLDLR and ApoER2 triggers phosphorylation of the cytoplasmic adaptor protein disabled 1 (Dab1) by Src family kinases, in particular Fyn. This series of reactions is required for cortical layer formation and for hippocampal dendrite development [73]. In addition, cyclin-dependent kinase 5-dependent signals are required for the function of reelin not only in neuronal migration but also in synaptic transmission [74]. The reelin signaling pathway is also involved in modulating learning and behavior [75]. Reelin can regulate NMDA-type glutamate receptor activity and potentiate calcium influx through NMDA receptors in neuronal cultures [75]. Sinagra et al. [76] documented a reelin-controlled change in subunit composition of NMDA glutamate receptors during maturation. Reelin signaling through ApoE receptors plays an essential role in synaptic plasticity and function in the adult brain [77].
Interestingly, BDNF, brain-derived neurotrophic factor, a primary target of MeCP2 [78, 79] regulates reelin production in cortical neurons. Bdnf -/- mice have elevated reelin levels in Cajal-Retzius cells. Overexpression of BDNF produces brain abnormalities similar to the phenotype of reeler (rl) mutant mice. This phenotype is preserved in slice cultures of hippocampus [80]. Furthermore, in in vitro co-culture systems, exogenous reelin caused dispersal of chain-migrating interneuron precursors in the olfactory bulb, suggesting that overexpression of reelin may affect neuronal migration [81].
We hypothesize that Reln overexpression is responsible for part of the phenotype of MeCP2-deficient mice, and that this phenotype is caused by abnormal neuronal migration in the developing brain of MeCP2-deficient mice and by dysregulation of reelin signaling pathways, thus disturbing synaptic function, in postnatal brain. To test this hypothesis, we have initiated genetic interaction studies. We aim to reduce the level of Reln expression in the Mecp2 mutant background by crossing rl+/- heterozygous males to Mecp2+/- female mice. We have preliminary evidence that the onset of morbid phenotypes is delayed and lifespan prolonged in some of the Mecp2Y/- ;Rln+/- double-mutant mice, when compared to their Mecp2Y/- ;Rln+/+ littermates.
Gtl2 (gene trap locus 2)
By microarray analysis, we identified Gtl2, called MEG3 (Maternally-expressed gene 3) in humans, as a significantly increased transcript in mutant cerebellum samples. qRT-PCR confirmed increased expression, although not significant due to large biological variation in different litters, in cerebellum and forebrain. Gtl2 encodes a maternally expressed imprinted non-translated RNA of unknown function. Gtl2 is expressed in most tissues, but most highly in brain. It lies within a cluster of imprinted genes on chromosome 12 that is conserved on distal 14q in human. Alternatively-spliced Gtl2 transcripts extend to include intron-encoded C/D box snoRNAs [82] and microRNAs [83, 84]. Thus, Gtl2 might function as a host gene for these small RNAs. In our qRT-PCR assays, the increase in Gtl2 expression in cerebellum was less striking, but Gtl2 expression was consistently increased in the forebrain of Mecp2-deficient mice.
Two previous studies have identified Gtl2 as a possible target of MeCP2. An expression microarray study on whole brain of one B-allele mouse revealed a 1.9-fold increase in Gtl2 expression [25]. Independently, by using a variant of differential display technology and qRT-PCR confirmation, Kriaucionis and colleagues [27] detected increased Gtl2 expression in whole brains of B-allele mice. Since the increase was significant only at late symptomatic stages, the authors suggested that it may represent a secondary consequence of the disease pathology. In contrast, our data show a similar increase in Gtl2 expression at 2 wk and at 8 wk, albeit detected by two different cDNA clones on the array. Gtl2 has a CpG-rich promoter region that is unmethylated on the maternal allele and becomes hypermethylated on the paternal allele after fertilization [85, 86]. By ChIP assay, we found that MeCP2 binds in vivo to this region with binding sites extending from 5' of exon 1 into intron 1, raising the possibility that it may play a role in silencing of the paternal allele. Alternatively, MeCP2 may bind to methylated CpG sites on the active maternal chromosome and thus regulate Gtl2 repression in certain brain regions or cell types. Future studies of allele-specific expression levels will address these possibilities.
Considering other previously reported candidate MeCP2 target genes that are represented on our microarrays: Only Snrp70, reported to be increased by qRT-PCR in whole brain from late symptomatic B-allele mice (1.44 fold, p = 0.04)[27], showed increased expression (1.5-fold) in cerebellum of 8 wk B-allele mice in our study. None of the following genes had significant expression changes in cerebellum: Uqcrc1 [27], Bdnf [87], Dlx5 [4], Fkbp5 and Sgk1 [25]. This is not surprising given that some of these genes may be primarily expressed in other brain regions and/or only in certain neuronal sub-types. Or transcript levels may be variable, when expression is modulated by activity state as for Bdnf [78, 79]. Our experience with biological validation and hierarchical clustering of independent samples taught us about the mitigating effects of different sibship identities. We need to consider that the different mothers are Mecp2+/- heterozygotes, having the genotype of females with RTT. Although not obviously symptomatic yet, their variable X-inactivation status could influence expression profiles in their offspring.
Conclusion
Cerebellar gene expression profiling of two Mecp2-mutant mouse models at three postnatal time points revealed no significant global changes. Irak1, located close to Mecp2, showed the most significant expression increase, but this may be specific to the large deletion in B-allele mice. We identified three novel direct MeCP2 target genes by increased expression and binding of MeCP2 to their promoters. Variable results in different litters do no preclude the potential importance of these genes. To evaluate the contributions of these mis-regulated genes to the synaptic dysfunction and neuropathologic phenotype observed in MeCP2-deficient humans and mice, further studies using higher cytological resolution, laser-capture dissection and immunohistochemical assays, as well as genetic interaction studies and functional assays of the affected pathways, will be needed.
Abbreviations
- RTT:
-
Rett syndrome
- MeCP2:
-
methyl-CpG binding protein 2
- DEG:
-
differentially expressed genes
- ChIP:
-
chromatin immunoprecipitation
References
Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A: Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992, 69 (6): 905-914. 10.1016/0092-8674(92)90610-O.
Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I, Bird AP: DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol Cell. 2005, 19 (5): 667-678. 10.1016/j.molcel.2005.07.021.
Koch C, Stratling WH: DNA binding of methyl-CpG-binding protein MeCP2 in human MCF7 cells. Biochemistry. 2004, 43 (17): 5011-5021. 10.1021/bi0359271.
Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T: Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet. 2005, 37 (1): 31-40.
Harikrishnan KN, Chow MZ, Baker EK, Pal S, Bassal S, Brasacchio D, Wang L, Craig JM, Jones PL, Sif S, El-Osta A: Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat Genet. 2005, 37 (3): 254-264. 10.1038/ng1516.
Drewell RA, Goddard CJ, Thomas JO, Surani MA: Methylation-dependent silencing at the H19 imprinting control region by MeCP2. Nucleic Acids Res. 2002, 30 (5): 1139-1144. 10.1093/nar/30.5.1139.
Muller C, Readhead C, Diederichs S, Idos G, Yang R, Tidow N, Serve H, Berdel WE, Koeffler HP: Methylation of the cyclin A1 promoter correlates with gene silencing in somatic cell lines, while tissue-specific expression of cyclin A1 is methylation independent. Mol Cell Biol. 2000, 20 (9): 3316-3329. 10.1128/MCB.20.9.3316-3329.2000.
Magdinier F, Billard LM, Wittmann G, Frappart L, Benchaib M, Lenoir GM, Guerin JF, Dante R: Regional methylation of the 5' end CpG island of BRCA1 is associated with reduced gene expression in human somatic cells. Faseb J. 2000, 14 (11): 1585-1594. 10.1096/fj.14.11.1585.
Georgel PT, Horowitz-Scherer RA, Adkins N, Woodcock CL, Wade PA, Hansen JC: Chromatin compaction by human MeCP2. Assembly of novel secondary chromatin structures in the absence of DNA methylation. J Biol Chem. 2003, 278 (34): 32181-32188. 10.1074/jbc.M305308200.
Nikitina T, Shi X, Ghosh RP, Horowitz-Scherer RA, Hansen JC, Woodcock CL: Multiple modes of interaction between the methylated DNA binding protein MeCP2 and chromatin. Mol Cell Biol. 2006, 27: 864-877. 10.1128/MCB.01593-06.
Young JI, Hong EP, Castle JC, Crespo-Barreto J, Bowman AB, Rose MF, Kang D, Richman R, Johnson JM, Berget S, Zoghbi HY: Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc Natl Acad Sci U S A. 2005, 102 (49): 17551-17558. 10.1073/pnas.0507856102.
Brero A, Easwaran HP, Nowak D, Grunewald I, Cremer T, Leonhardt H, Cardoso MC: Methyl CpG-binding proteins induce large-scale chromatin reorganization during terminal differentiation. J Cell Biol. 2005, 169 (5): 733-743. 10.1083/jcb.200502062.
Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY: Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999, 23: 185-188. 10.1038/13810.
Wan M, Lee SS, Zhang X, Houwink-Manville I, Song HR, Amir RE, Budden S, Naidu S, Pereira JL, Lo IF, Zoghbi HY, Schanen NC, Francke U: Rett syndrome and beyond: recurrent spontaneous and familial MECP2 mutations at CpG hotpots. Am J Hum Genet. 1999, 65 (6): 1520-1529. 10.1086/302690.
Archer HL, Whatley SD, Evans JC, Ravine D, Huppke P, Bunyan D, Kerr AM, Kerr B, Sweeney E, Davies SJ, Reardon W, Horn J, Macdermot KD, Smith RA, Magee A, Donaldson A, Crow Y, Hermon G, Miedzybrodzka Z, Cooper DN, Lazarou L, Butler R, Sampson JR, Pilz DT, Laccone F, Clarke AJ: Gross rearrangements of the MECP2 gene are found in both classical and atypical Rett Syndrome. J Med Genet. 2006, 43: 451-456. 10.1136/jmg.2005.033464.
Rett A: Uber ein eigenartiges hirnatrophisches Syndrom bei Hyperammonamie im Kindesalter. Wien Med Wochenschr. 1966, 116: 723-738.
Hagberg B, Aicardi J, Dias K, Ramos O: A progressive syndrome of autism, dementia, ataxia, and loss of purposeful hand use in girls: Rett's syndrome: report of 35 cases. Ann Neurol. 1983, 14 (4): 471-479. 10.1002/ana.410140412.
Glaze DG: Rett syndrome: of girls and mice--lessons for regression in autism. Ment Retard Dev Disabil Res Rev. 2004, 10 (2): 154-158. 10.1002/mrdd.20030.
Francke U: Neurogenetics of MeCP2 deficiency. Nature Clinical Practice Neurology. 2006, 2 (April): 212-221. 10.1038/ncpneuro0148.
Kankirawatana P, Leonard H, Ellaway C, Scurlock J, Mansour A, Makris CM, Dure LS, Friez M, Lane J, Kiraly-Borri C, Fabian V, Davis M, Jackson J, Christodoulou J, Kaufmann WE, Ravine D, Percy AK: Early progressive encephalopathy in boys and MECP2 mutations. Neurology. 2006, 67 (1): 164-166. 10.1212/01.wnl.0000223318.28938.45.
Colantuoni C, Jeon OH, Hyder K, Chenchik A, Khimani AH, Narayanan V, Hoffman EP, Kaufmann WE, Naidu S, Pevsner J: Gene expression profiling in postmortem Rett Syndrome brain: differential gene expression and patient classification. Neurobiol Dis. 2001, 8 (5): 847-865. 10.1006/nbdi.2001.0428.
Traynor J, Agarwal P, Lazzeroni L, Francke U: Gene expression patterns vary in clonal cell cultures from Rett syndrome females with eight different MECP2 mutations. BMC Med Genet. 2002, 3 (1): 12-10.1186/1471-2350-3-12.
Tudor M, Akbarian S, Chen RZ, Jaenisch R: Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes in the brain. Proc Natl Acad Sci U S A. 2002, 99 (24): 15536-15541. 10.1073/pnas.242566899.
Ballestar E, Ropero S, Alaminos M, Armstrong J, Setien F, Agrelo R, Fraga MF, Herranz M, Avila S, Pineda M, Monros E, Esteller M: The impact of MECP2 mutations in the expression patterns of Rett syndrome patients. Hum Genet. 2005, 116 (1-2): 91-104. 10.1007/s00439-004-1200-0.
Nuber UA, Kriaucionis S, Roloff TC, Guy J, Selfridge J, Steinhoff C, Schulz R, Lipkowitz B, Ropers HH, Holmes MC, Bird A: Up-regulation of glucocorticoid-regulated genes in a mouse model of Rett syndrome. Hum Mol Genet. 2005, 14 (15): 2247-2256. 10.1093/hmg/ddi229.
Delgado IJ, Kim DS, Thatcher KN, LaSalle JM, Van den Veyver IB: Expression profiling of clonal lymphocyte cell cultures from Rett syndrome patients. BMC Med Genet. 2006, 7: 61-10.1186/1471-2350-7-61.
Kriaucionis S, Paterson A, Curtis J, Guy J, Macleod N, Bird A: Gene expression analysis exposes mitochondrial abnormalities in a mouse model of Rett syndrome. Mol Cell Biol. 2006, 26 (13): 5033-5042. 10.1128/MCB.01665-05.
Nadler JJ, Zou F, Huang H, Moy SS, Lauder J, Crawley JN, Threadgill DW, Wright FA, Magnuson TR: Large-scale gene expression differences across brain regions and inbred strains correlate with a behavioral phenotype. Genetics. 2006, 174 (3): 1229-1236. 10.1534/genetics.106.061481.
Mullaney BC, Johnston MV, Blue ME: Developmental expression of methyl-CpG binding protein 2 is dynamically regulated in the rodent brain. Neuroscience. 2004, 123 (4): 939-949. 10.1016/j.neuroscience.2003.11.025.
Chen RZ, Akbarian S, Tudor M, Jaenisch R: Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001, 27 (3): 327-331. 10.1038/85906.
Guy J, Hendrich B, Holmes M, Martin JE, Bird A: A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001, 27 (3): 322-326. 10.1038/85899.
Oldfors A, Sourander P, Armstrong DL, Percy AK, Witt-Engerstrom I, Hagberg BA: Rett syndrome: cerebellar pathology. Pediatr Neurol. 1990, 6 (5): 310-314. 10.1016/0887-8994(90)90022-S.
Jordan C, Francke U: Ube3a expression is not altered in Mecp2 mutant mice. Hum Mol Genet. 2006, 15 (14): 2210-2215. 10.1093/hmg/ddl146.
SFGF: Stanford Functional Genomics Facility. [http://www.microarray.org/sfgf/]
SOURCE: SOURCE. [http://source.stanford.edu]
Brown: Brown Lab Experimental Protocols. [http://cmgm.stanford.edu/pbrown/protocols]
Tusher VG, Tibshirani R, Chu G: Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001, 98 (9): 5116-5121. 10.1073/pnas.091062498.
Eisen MB, Spellman PT, Brown PO, Botstein D: Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998, 95 (25): 14863-14868. 10.1073/pnas.95.25.14863.
Sherlock G, Hernandez-Boussard T, Kasarskis A, Binkley G, Matese JC, Dwight SS, Kaloper M, Weng S, Jin H, Ball CA, Eisen MB, Spellman PT, Brown PO, Botstein D, Cherry JM: The Stanford Microarray Database. Nucleic Acids Res. 2001, 29 (1): 152-155. 10.1093/nar/29.1.152.
Oberley MJ, Farnham PJ: Probing chromatin immunoprecipitates with CpG-island microarrays to identify genomic sites occupied by DNA-binding proteins. Methods Enzymol. 2003, 371: 577-596.
Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Bass MB, Arruda I, Robinson MO: A mammalian telomerase-associated protein. Science. 1997, 275 (5302): 973-977. 10.1126/science.275.5302.973.
Kickhoefer VA, Stephen AG, Harrington L, Robinson MO, Rome LH: Vaults and telomerase share a common subunit, TEP1. J Biol Chem. 1999, 274 (46): 32712-32717. 10.1074/jbc.274.46.32712.
Kickhoefer VA, Liu Y, Kong LB, Snow BE, Stewart PL, Harrington L, Rome LH: The Telomerase/vault-associated protein TEP1 is required for vault RNA stability and its association with the vault particle. J Cell Biol. 2001, 152 (1): 157-164. 10.1083/jcb.152.1.157.
Schmidt JV, Matteson PG, Jones BK, Guan XJ, Tilghman SM: The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes Dev. 2000, 14 (16): 1997-2002.
Wade PA, Jones PL, Vermaak D, Veenstra GJ, Imhof A, Sera T, Tse C, Ge H, Shi YB, Hansen JC, Wolffe AP: Histone deacetylase directs the dominant silencing of transcription in chromatin: association with MeCP2 and the Mi-2 chromodomain SWI/SNF ATPase. Cold Spring Harb Symp Quant Biol. 1998, 63: 435-445. 10.1101/sqb.1998.63.435.
Hu K, Nan X, Bird A, Wang W: Testing for association between MeCP2 and the brahma-associated SWI/SNF chromatin-remodeling complex. Nat Genet. 2006, 38 (9): 962-4; author reply 964-7. 10.1038/ng0906-962.
Dickinson LA, Joh T, Kohwi Y, Kohwi-Shigematsu T: A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell. 1992, 70 (4): 631-645. 10.1016/0092-8674(92)90432-C.
Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T: SATB1 targets chromatin remodelling to regulate genes over long distances. Nature. 2002, 419 (6907): 641-645. 10.1038/nature01084.
Cai S, Lee CC, Kohwi-Shigematsu T: SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006, 38 (11): 1278-1288. 10.1038/ng1913.
Britanova O, Akopov S, Lukyanov S, Gruss P, Tarabykin V: Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. Eur J Neurosci. 2005, 21 (3): 658-668. 10.1111/j.1460-9568.2005.03897.x.
von Kries JP, Buhrmester H, Stratling WH: A matrix/scaffold attachment region binding protein: identification, purification, and mode of binding. Cell. 1991, 64 (1): 123-135. 10.1016/0092-8674(91)90214-J.
Croteau S, Charron MC, Latham KE, Naumova AK: Alternative splicing and imprinting control of the Meg3/Gtl2-Dlk1 locus in mouse embryos. Mamm Genome. 2003, 14 (4): 231-241. 10.1007/s00335-002-2244-x.
Deng V, Matagne V, Banine F, Frerking M, Ohliger P, Budden S, Pevsner J, Dissen GA, Sherman LS, Ojeda SR: FXYD1 is a MeCP2 target gene overexpressed in the brains of Rett syndrome patients and Mecp2-null mice. Hum Mol Genet. 2007, 16 (6): 640-650. 10.1093/hmg/ddm007.
Luikenhuis S, Giacometti E, Beard CF, Jaenisch R: Expression of MeCP2 in postmitotic neurons rescues Rett syndrome in mice. Proc Natl Acad Sci U S A. 2004, 101 (16): 6033-6038. 10.1073/pnas.0401626101.
Braunschweig D, Simcox T, Samaco RC, LaSalle JM: X-Chromosome inactivation ratios affect wild-type MeCP2 expression within mosaic Rett syndrome and Mecp2-/+ mouse brain. Hum Mol Genet. 2004, 13 (12): 1275-1286. 10.1093/hmg/ddh142.
Su J, Richter K, Zhang C, Gu Q, Li L: Differential regulation of interleukin-1 receptor associated kinase 1 (IRAK1) splice variants. Mol Immunol. 2007, 44 (5): 900-905. 10.1016/j.molimm.2006.03.021.
Geering K: FXYD proteins: new regulators of Na-K-ATPase. Am J Physiol Renal Physiol. 2006, 290 (2): F241-50. 10.1152/ajprenal.00126.2005.
Feschenko MS, Donnet C, Wetzel RK, Asinovski NK, Jones LR, Sweadner KJ: Phospholemman, a single-span membrane protein, is an accessory protein of Na,K-ATPase in cerebellum and choroid plexus. J Neurosci. 2003, 23 (6): 2161-2169.
Beevers AJ, Kukol A: Secondary structure, orientation, and oligomerization of phospholemman, a cardiac transmembrane protein. Protein Sci. 2006, 15 (5): 1127-1132. 10.1110/ps.051899406.
Garty H, Karlish SJ: Role of FXYD proteins in ion transport. Annu Rev Physiol. 2006, 68: 431-459. 10.1146/annurev.physiol.68.040104.131852.
Ahlers BA, Zhang XQ, Moorman JR, Rothblum LI, Carl LL, Song J, Wang J, Geddis LM, Tucker AL, Mounsey JP, Cheung JY: Identification of an endogenous inhibitor of the cardiac Na+/Ca2+ exchanger, phospholemman. J Biol Chem. 2005, 280 (20): 19875-19882. 10.1074/jbc.M414703200.
Jia LG, Donnet C, Bogaev RC, Blatt RJ, McKinney CE, Day KH, Berr SS, Jones LR, Moorman JR, Sweadner KJ, Tucker AL: Hypertrophy, increased ejection fraction, and reduced Na-K-ATPase activity in phospholemman-deficient mice. Am J Physiol Heart Circ Physiol. 2005, 288 (4): H1982-8. 10.1152/ajpheart.00142.2004.
Jespersen T, Grunnet M, Rasmussen HB, Jorgensen NB, Jensen HS, Angelo K, Olesen SP, Klaerke DA: The corticosteroid hormone induced factor: a new modulator of KCNQ1 channels?. Biochem Biophys Res Commun. 2006, 341 (4): 979-988. 10.1016/j.bbrc.2006.01.046.
McGill BE, Bundle SF, Yaylaoglu MB, Carson JP, Thaller C, Zoghbi HY: Enhanced anxiety and stress-induced corticosterone release are associated with increased Crh expression in a mouse model of Rett syndrome. Proc Natl Acad Sci U S A. 2006, 103 (48): 18267-18272. 10.1073/pnas.0608702103.
Chen Y, Sharma RP, Costa RH, Costa E, Grayson DR: On the epigenetic regulation of the human reelin promoter. Nucleic Acids Res. 2002, 30 (13): 2930-2939. 10.1093/nar/gkf401.
Dong E, Agis-Balboa RC, Simonini MV, Grayson DR, Costa E, Guidotti A: Reelin and glutamic acid decarboxylase67 promoter remodeling in an epigenetic methionine-induced mouse model of schizophrenia. Proc Natl Acad Sci U S A. 2005, 102 (35): 12578-12583. 10.1073/pnas.0505394102.
Dong E, Guidotti A, Grayson DR, Costa E: Histone hyperacetylation induces demethylation of reelin and 67-kDa glutamic acid decarboxylase promoters. Proc Natl Acad Sci U S A. 2007, 104 (11): 4676-4681. 10.1073/pnas.0700529104.
Tissir F, Goffinet AM: Reelin and brain development. Nat Rev Neurosci. 2003, 4 (6): 496-505. 10.1038/nrn1113.
Hatten ME: Central nervous system neuronal migration. Annu Rev Neurosci. 1999, 22: 511-539. 10.1146/annurev.neuro.22.1.511.
Alcantara S, Ruiz M, D'Arcangelo G, Ezan F, de Lecea L, Curran T, Sotelo C, Soriano E: Regional and cellular patterns of reelin mRNA expression in the forebrain of the developing and adult mouse. J Neurosci. 1998, 18 (19): 7779-7799.
Weeber EJ, Beffert U, Jones C, Christian JM, Forster E, Sweatt JD, Herz J: Reelin and ApoE receptors cooperate to enhance hippocampal synaptic plasticity and learning. J Biol Chem. 2002, 277 (42): 39944-39952. 10.1074/jbc.M205147200.
Trommsdorf M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, Nimpf J, Hammer RE, Richardson JA, Herz J: Reeler/disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell. 1999, 97 (6): 689-701. 10.1016/S0092-8674(00)80782-5.
Howell BW, Herrick TM, Hildebrand JD, Zhang Y, Cooper JA: Dab1 tyrosine phosphorylation sites relay positional signals during mouse brain development. Curr Biol. 2000, 10 (15): 877-885. 10.1016/S0960-9822(00)00608-4.
Beffert U, Weeber EJ, Morfini G, Ko J, Brady ST, Tsai LH, Sweatt JD, Herz J: Reelin and cyclin-dependent kinase 5-dependent signals cooperate in regulating neuronal migration and synaptic transmission. J Neurosci. 2004, 24 (8): 1897-1906. 10.1523/JNEUROSCI.4084-03.2004.
Chen Y, Beffert U, Ertunc M, Tang TS, Kavalali ET, Bezprozvanny I, Herz J: Reelin modulates NMDA receptor activity in cortical neurons. J Neurosci. 2005, 25 (36): 8209-8216. 10.1523/JNEUROSCI.1951-05.2005.
Sinagra M, Verrier D, Frankova D, Korwek KM, Blahos J, Weeber EJ, Manzoni OJ, Chavis P: Reelin, very-low-density lipoprotein receptor, and apolipoprotein E receptor 2 control somatic NMDA receptor composition during hippocampal maturation in vitro. J Neurosci. 2005, 25 (26): 6127-6136. 10.1523/JNEUROSCI.1757-05.2005.
Herz J, Chen Y: Reelin, lipoprotein receptors and synaptic plasticity. Nat Rev Neurosci. 2006, 7 (11): 850-859. 10.1038/nrn2009.
Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME: Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003, 302 (5646): 885-889. 10.1126/science.1086446.
Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE: DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003, 302 (5646): 890-893. 10.1126/science.1090842.
Zhao S, Chai X, Forster E, Frotscher M: Reelin is a positional signal for the lamination of dentate granule cells. Development. 2004, 131: 5117-5125. 10.1242/dev.01387.
Hack I, Bancila M, Loulier K, Carroll P, Cremer H: Reelin is a detachment signal in tangential chain-migration during postnatal neurogenesis. Nat Neurosci. 2002, 5 (10): 939-945. 10.1038/nn923.
Cavaille J, Seitz H, Paulsen M, Ferguson-Smith AC, Bachellerie JP: Identification of tandemly-repeated C/D snoRNA genes at the imprinted human 14q32 domain reminiscent of those at the Prader-Willi/Angelman syndrome region. Hum Mol Genet. 2002, 11 (13): 1527-1538. 10.1093/hmg/11.13.1527.
Lin SP, Youngson N, Takada S, Seitz H, Reik W, Paulsen M, Cavaille J, Ferguson-Smith AC: Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet. 2003, 35 (1): 97-102. 10.1038/ng1233.
Tierling S, Dalbert S, Schoppenhorst S, Tsai CE, Oliger S, Ferguson-Smith AC, Paulsen M, Walter J: High-resolution map and imprinting analysis of the Gtl2-Dnchc1 domain on mouse chromosome 12. Genomics. 2006, 87 (2): 225-235. 10.1016/j.ygeno.2005.09.018.
Miyoshi N, Wagatsuma H, Wakana S, Shiroishi T, Nomura M, Aisaka K, Kohda T, Surani MA, Kaneko-Ishino T, Ishino F: Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells. 2000, 5 (3): 211-220. 10.1046/j.1365-2443.2000.00320.x.
Takada S, Tevendale M, Baker J, Georgiades P, Campbell E, Freeman T, Johnson MH, Paulsen M, Ferguson-Smith AC: Delta-like and gtl2 are reciprocally expressed, differentially methylated linked imprinted genes on mouse chromosome 12. Curr Biol. 2000, 10 (18): 1135-1138. 10.1016/S0960-9822(00)00704-1.
Chang Q, Khare GD, Dani VS, Nelson S, Jaenisch R: The disease progression of Mecp2 mutant mice is affected by the level of BDNF expression. Neuron. 2006, 49: 341-348. 10.1016/j.neuron.2005.12.027.
Stanford Microarray Database. [http://smd.stanford.edu/cgi-bin/publication/viewPublication.pl?pub_no=630]
EBI ArrayExpress. [http://www.ebi.ac.uk/arrayexpress/#ae-browse/q=E-SMDB-2451]
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2350/8/36/prepub
Acknowledgements
The authors are grateful to Francke Lab members for helpful discussions and assistance, to Kate Rubins for helping with microarrays, to Craig April and Jon Pollack for advice in designing experiments and analyzing data, to Birgitt Schüle for human brain RNA samples, and to Peggy Farnham for advice on ChIP protocols.
Funding: C.J. was supported by the Stanford Genome Training Program (T32 HG00044 from the National Human Genome Research Institute). This work was supported in part by research grants from the NIH and the International Rett Syndrome Association.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
CJ carried out the microrray and qRT-PCR studies, and the statistical analyses, and drafted the manuscript. HHL carried out the chromatin immunoprecipitation assays, validation studies and qRT-PCR of human brain samples. HCK participated in mouse breeding, genotyping and experimental design. UF conceived of the study, participated in its design, interpreted the results and prepared the final manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Jordan, C., Li, H.H., Kwan, H.C. et al. Cerebellar gene expression profiles of mouse models for Rett syndrome reveal novel MeCP2 targets. BMC Med Genet 8, 36 (2007). https://doi.org/10.1186/1471-2350-8-36
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2350-8-36