Abstract
Background
Multinodular goitre (MNG) is a common disorder characterised by an enlargement of the thyroid, occurring as a compensatory response to hormonogenesis impairment. The incidence of MNG is dependent on sex (female:male ratio 5:1) and several reports have documented a genetic basis for the disease. Last year we mapped a MNG locus to chromosome Xp22 in a region containing the peroxiredoxin IV (Prx-IV) gene. Since Prx-IV is involved in the removal of H2O2 in thyroid cells, we hypothesize that mutations in Prx-IV gene are involved in pathogenesis of MNG.
Methods
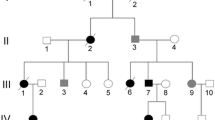
Four individuals (2 affected, 2 unrelated unaffected) were sequenced using automated methods. All individuals were originated from the original three-generation Italian family described in previous studies. A Southern blot analysis using a Prx-IV full-length cDNA as a probe was performed in order to exclude genomic rearrangements and/or intronic mutations. In addition a RT-PCR of PRX-IV was performed in order to investigate expression alterations.
Results
No causative mutations were found. Two adjacent nucleotide substitutions were detected within introns 1 and 4. These changes were also detected in unaffected individuals, suggesting that they were innocuous polymorphisms. No gross genomic rearrangements and/or restriction fragment alterations were observed on Southern analysis. Finally, using RT-PCR from tissue-specific RNA, no differences of PRX-IV expression-levels were detected between affected and unaffected samples.
Conclusions
Based on sequence and genomic analysis, Prx-IV is very unlikely to be the MNG2 gene.
Similar content being viewed by others
Background
Multinodular goiter (MNG, OMIM *138800) is a common disorder, arising as a compensatory response to an impairment of thyroid hormonogenesis. Affected individuals maintain normal hormone levels by an increase in thyroid activity and mass, so that they are eu-metabolic but goitrous [1]. Even if MNG incidence is influenced by iodine intake, family clustering of the disease has been reported in iodo-sufficient areas, suggesting a genetic basis for the disease [2]. This was confirmed by the identification of three dominant MNG loci: MNG1 on chromosome 14q [3], MNG2 on chromosome Xp22 [4] and MNG3 on chromosome 3q26 [5]. The MNG2 locus maps to a 9.6 cM interval containing the peroxiredoxin-IV (Prx-IV, formerly known as AOE372) gene. Peroxiredoxins are a family of highly conserved anti-oxidant enzymes, reducing hydrogen peroxide and/or organic hydroperoxides [6]. In thyroid cells, regulation of H2O2 concentration is critical for the thyroxine synthesis, since hydrogen peroxide is needed for the conversion of iodide to organically bound iodine. Since PRX-I e PRX-II are involved in the removal of H2O2 in thyroid cells, we have hypothesised that Prx-IV mutations may affect intracellular H2O2 levels and impair thyroxine hormonogenesis. Therefore, we have screened Prx-IV as a MNG2 positional candidate.
Methods
Direct sequencing
We have sequenziated four individuals (2 affected, 2 unrelated unaffected) originated from the large family that allowed the MNG2 locus assignment [4].
Intronic primers pairs amplifying Prx-IV exons were designed (Table 1), based on the genomic structure reported by Wong et al.[6]. One primers pair amplifying 500 bp of putative promoter was also selected (Table 1). Intron and 5'UTR sequences were obtained, using the BLAST program to match Prx-IV mRNA (accession n. XM_041446) against the "htgs" database of genomic sequences. The most likely location of the promoter was assessed using the SCAN SIGNAL software. PCR products were sequenced with the CEQ™ DTCS Kit (Beckman-Coulter, CA, USA) and run on a CEQ2000 automated sequencer (Beckman-Coulter, CA, USA).
Southern blot
5 μg genomic DNA were digested using EcoRI and Hind III. Digests were electrophoresed on 1.0% agarose gels, and the DNA was subsequently transferred to Hybond N+ filter membranes (Amersham), with 10 × SSC as transferring buffer. Probes were labeled, by random priming, with 32P-dCTP. Membranes were hybridized overnight at 65°C. Final washing of the membranes was performed with 0.1 × SSC at 65°C. The results were analyzed with the STORM Phosphor Imaging System.
FNAB and RT-PCR
Patients were placed in the recumbent position with a pillow under their shoulders, such that their neck was hyperextended, and the skin cleansed with povidone iodine (Betadina scrub). The nodules was fixed in position manually, and a 23 gauge needle attacched to a 20 ml disposable syringe inserted perpendicular to the anterior surface of the neck. Once the needle was in place, costant suction was applied and maintained while the needle was withdrawn to the level of the nodules capsule. The content was then discharged onto glass slides and smears were made.
FNAB samples were then washed twice with 1 × PBS and then processed for RNA extraction. Approximately 1 μg of RNA was incubated at 42°C for 1 hour with 2.5 μM random hexamers (Pharmacia, LKB, Uppsala), 25 U of Rnase inhibitor (Rnasin, Promega, Madison, WI), 2.5 μl of 10 mM each dNTPs, 4 μl of 5 × AMV RT (Promega, Madison, WI) in a final volume of 20 μl. The cDNA was next amplified by PCR for PRX-IV gene using primers showed in table 1. As a positive control we coamplified the constitutively expressed enzyme glyceraldeide 3-phosphate dehydrogenase (GAPDH).
The intensity of the amplified fragments was compared by scanning using a Molecular Dynamics (Sunnyvale, CA) densitometer (ImageQuant).
Results
We have carried out Prx-IV mutational analysis in the three-generation Italian pedigree that allowed the MNG2 locus assignment [4]. Prx-IV is a cytoplasmatic thioredoxin peroxidase that detoxifies hydrogen peroxide in a redox chain leading to NADPH oxidation [7]. Therefore, Prx-IV appears an attractive MNG candidate, since thyroxine synthesis requires hydrogen peroxide.
Patient sequencing identified two adjacent substitutions in intron 1 (IVS1-9G>A and IVS1-10A>T), and one in intron 4 (IVS4+81T>C). All variants were also found in one unrelated unaffected male, thus ruling out the hypothesis that any of them might be disease-related. Coding region analysis failed to detect any additional substitutions and moreover promoter region analysis didn't detect other substitutions. Since the affected females carrying both alleles at intron 2 variants, the occurrence of a large gene deletion can be excluded. In addition, as shown in figure 1, no gross PRX-IV rearrangements were detected by Southern blot analysis.
Expression analysis using amplified PRX-IV cDNA revealed a single PCR product of the expected size. An equal relative ratio of intensity with a value 0.86 was detected when this product was compared with GAPDH cDNA used as a control (Data not shown).
Discussion and Conclusions
Altogether our data indicate that Prx-IV is very unlikely to be the MNG2 gene.
The MNG2 region does not contain any additional obvious candidate, but many ESTs map to the critical interval. Thus, our future research will be devoted to the identification and characterization of those MNG2 ESTs that are expressed in the thyroid.
Abbreviations
- MNG:
-
multinodular goitre
- PRX:
-
peroxiredoxin
- EST:
-
expressed sequence tag
- FNAB:
-
fine needle aspiration biopsy
References
Ingbar SH, Woeber KA: The thyroid gland. In: Textbook of endocrinology. Edited by: Williams RH. 1968, WB Saunders, Philadelphia, 105-279.
Greig WR, Boyle JA, Duncan A: Genetic and non-genetic factors in simple goitre formation: evidence from a twin study. Q J Med. 1967, 36: 175-188.
Bignell GR, Canzian F, Shayeghi M, Stark M, Shugart YY, Biggs P, Mangion J, Hamoudi R, Rosenblat J, Buu P, Sun S, Stoffer SS, Goldgar DE, Romeo G, Houlston RS, Narod SA, Stratton MR, Foulkes WD: Familial non toxic multinodular goitre locus maps to chromosome 14q but does not account for familial non medullary thyroid cancer. Am J Hum Genet. 1997, 61: 1123-1130. 10.1086/301610.
Capon F, Tacconelli A, Giardina E, Sciacchitano S, Bruno R, Tassi V, Trsichitta V, Filetti S, Dallapiccola B, Novelli G: Mapping a dominant form of multinodular goitre to chromosome Xp22. Am J Hum Genet. 2000, 67: 1004-1007. 10.1086/303095.
Takahashi T, Nozaki J, Komatsu M, Wada Y, Utsunomiya M, Inoue K, Takada G, Koizumi A: A new locus for a dominant form of Multinodular goiter on 3q26.1-q26.3. Biochem Biophys Res Commun. 2001, 284: 650-654. 10.1006/bbrc.2001.4998.
Wong CM, Chun ACS, Kok KH, Zhou Y, Fung PCW, Kung HF, Jeang KT, Jin DY: Characterisation of human and mouse peroxiredoxin IV: evidence of inhibition by Prx-IV of epidermal growth factor- and p53-induced reactive oxygen species. Antioxid Redox Signal. 2000, 2: 507-518. 10.1089/15230860050192288.
Jin DY, Chae HZ, Rhee SG, Jeang KT: Regulatory role for a novel human thioredoxin peroxidase in NF-κB activation. J Biol Chem. 1997, 272: 30952-30961. 10.1074/jbc.272.49.30952.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2350/3/5/prepub
Acknowledgements
This work was funded by the Italian Telethon (project E1031).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
None declared
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Giardina, E., Capon, F., D'Apice, M.R. et al. Mutational analysis of Peroxiredoxin IV: exclusion of a positional candidate for multinodular goitre. BMC Med Genet 3, 5 (2002). https://doi.org/10.1186/1471-2350-3-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2350-3-5