Abstract
Background
Auditory neuropathy spectrum disorder (ANSD) is a unique form of hearing loss that involves absence or severe abnormality of auditory brainstem response (ABR), but also the presence of otoacoustic emissions (OAEs). However, with age, the OAEs disappear, making it difficult to distinguish this condition from other nonsyndromic hearing loss. Therefore, the frequency of ANSD may be underestimated. The aim of this study was to determine what portion of nonsyndromic hearing loss is caused by mutations of OTOF, the major responsible gene for nonsyndromic ANSD.
Methods
We screened 160 unrelated Japanese with severe to profound recessive nonsyndromic hearing loss (ARNSHL) without GJB2 or SLC26A4 mutations, and 192 controls with normal hearing.
Results
We identified five pathogenic OTOF mutations (p.D398E, p.Y474X, p.N727S, p.R1856Q and p.R1939Q) and six novel, possibly pathogenic variants (p.D450E, p.W717X, p.S1368X, p.R1583H, p.V1778I, and p.E1803A).
Conclusions
The present study showed that OTOF mutations accounted for 3.2–7.3% of severe to profound ARNSHL patients in Japan. OTOF mutations are thus a frequent cause in the Japanese deafness population and mutation screening should be considered regardless of the presence/absence of OAEs.
Similar content being viewed by others
Background
Auditory neuropathy (AN), a unique form of hearing loss, involves absence or severe abnormality of auditory brainstem response (ABR), but presence of otoacoustic emissions (OAE) and/or cochlear microphonic (CM). This disorder was defined by Starr [1], and also reported as “Auditory nerve disease” [2] and “Auditory dys-synchrony” [3]. AN was renamed “auditory neuropathy spectrum disorder (ANSD)” in 2008, due to the heterogeneous and multifaceted nature [4].
The prevalence of ANSD in sensorineural hearing loss is reported to be 0.5-15% [5]. The etiologies of ANSD are various; patients range from infants to adults, 42% of which are associated with hereditary neurological disorders, 10% with toxic, metabolic, immunological and infectious causes, and 48% with unknown causes [6]. Although the exact percentage of nonsyndromic ANSD is unclear, responsible genes have been gradually revealed. To date, mutations of AUNA1, OTOF, PJVK, GJB2 and mitochondrial 12S rRNA are reported to be causal for nonsyndromic ANSD [7].
The OTOF gene (DFNB9) is mainly expressed in cochlear inner hair cells, and is necessary for synaptic exocytosis at the auditory ribbon synapse [8]. It encodes both long and short isoforms with the long isoform containing six C2 domains and the C-terminal transmembrane domain, and the short isoform containing only the last three C2 domains [9]. Mutations in the OTOF gene, encoding otoferlin, are reported to be the major causes of nonsyndromic recessive ANSD [10–12]. In Japanese, mutations in OTOF account for 56. 5% (13/23) of ANSD [13]. Although ANSD can be characterized by the presence of OAEs in the first two years of life, OAEs later disappear and the hearing loss then resembles other types of nonsyndromic hearing loss [14]. Because of expected good outcomes of cochlear implantation for patients with OTOF mutations [15, 16], it is important to perform mutation screening for OTOF to select the appropriate intervention. Although some reports have described OTOF mutations in severe to profound autosomal recessive hearing loss patients in other populations [11, 12], there has been no literature available regarding the screening of OTOF mutations using a large cohort in a comprehensive manner. The goal of this study was therefore to reveal the frequency of ANSD and to identify OTOF mutations in Japanese ARNSHL patients.
Methods
Subjects
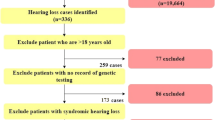
Among the 1511 Japanese independent hearing loss patients registered in our DNA sample bank, 469 were congenital severe to profound sensorineural hearing loss (above 71 dB average over 500, 1000, 2000 and 4000 Hz in the better hearing ear) patients compatible with autosomal recessive inheritance (including sporadic cases). From those, we randomly selected 160 patients. All ANSD cases were sporadic (compatible with autosomal recessive inheritance). They were diagnosed as ANSD by evaluation of OAE response. We excluded autosomal dominant families because in previous studies OTOF mutations were not found in such groups [17]. Pure tone audiometry was used for adults (N= 32) and ABR, auditory steady-state responses (ASSR), and conditioned orientation response audiometry (COR) were used for pediatric patients (n=128). The control group was composed of 192 unrelated Japanese individuals who had normal hearing shown by auditory testing. All subjects gave prior informed written consent for participation in the project and the Ethical Committee of Shinshu University approved the study.
Mutation analysis
We designed 43 pairs of primers to amplify DNA fragments containing all exons in the coding regions of the OTOF gene (ENST00000403946). Primer3Plus (http://www.bioinformatic.nl/cgi-bin/primer3plus/primer3plus.cgi) was used to design primers to flank all the exon-intron boundaries. Each genomic DNA sample (40 ng) was amplified, using Ampli Taq Gold (Applied Biosystems, Foster City, CA), for 5 min at 95°C, followed by 30 three-step cycles of 95°C for 30s, 60°C for 30s, and 72°C for 60s, with a final extension at 72°C for 7 min, ending with a holding period at 4°C in a PCR thermal cycler (Takara, Shiga, Japan). PCR products were treated with ExoSAP-IT® (GE Healthcare Bio, Santa Clara, CA) by incubation at 37°C for 60 min, and inactivation at 80°C for 15 min. After the products were purified, we performed standard cycle-sequencing reactions with ABI Big Dye® terminators in an ABI PRISM 3100 Genetic Analyzer autosequencer (Applied Biosystems, Foster City, CA).
Computer analysis to predict the effect of missense variants on the protein function was performed with wANNOVAR [18–20] (http://wannovar.usc.edu) including functional prediction software listed below. PhyloP (http://hgdownload.cse.ucsc.edu/goldenPath/hg18/phyloP44way/), Sorting Intolerant from Tolerant (SIFT; http://sift.jcvi.org/), Polymorphism Phenotyping (PolyPhen2; http://genetics.bwh.harvard.edu/pph2/), LRT (http://www.genetics.wustl.edu/jflab/lrt_query.html), and MutationTaster (http://www.mutationtaster.org/).
Results
We found a total of 11 probable pathogenic variants in the patients (Table 1). Among them, five mutations were previously reported: p.D398E, p.Y474X, p.N727S, p.R1856Q and p.R1939Q. The other six probable pathogenic variants were novel: 2 nonsense mutations (p.W717X, p.S1368X) and 4 missense mutations (p.D450E, p.R1583H, p.V1778I, p. E1803A). Based on the prediction programs, it is most likely that p.D450E (c.1350C>G), p.R1583H (c.4748G>A), p.V1778I (c.5332G>A), and p.E1803A (c.5408A>C) were pathogenic. In addition, they were absent (or in very few numbers) in the controls, and located in C2 domains, which are highly conserved among species (Figure 1). In addition, polymorphic changes were also identified (Table 2). p.R1676C (c.5026C>T) was previously reported to be pathogenic [21], but we excluded p.R1676C as it is unlikely to be pathologic because of high frequencies in the control population (Table 2). Among the 16 patients with OTOF mutations, 4 were homozygous, 3 were compound heterozygotes, and 9 were heterozygous without second mutation (Table 3). After clinical re-evaluation, we recategorized cases with OAE as ANSD.
The location of mutations in otoferlin protein and the evolutionary conservation of the amino acids. (A) Evolutionary conservation. The locations of mutations are boxed. (B) Novel pathogenic OTOF mutations found in this work and relation to the functional domains of otoferlin. C2A-F: C2 domains. TMD: transmembrane domain.
Discussion
So far, more than 90 pathologic mutations have been reported in OTOF[25]. The present study identified 11 possibly pathogenic OTOF variants in Japanese patients with nonsyndromic hearing loss, and 6 of them were novel mutations (p.D450E, p.W717X, p.S1368X, p.R1583H, p.V1778I, and p.E1803A). Concerning pathogenicity of the four novel missense mutations, p.R1583H is more likely to be a disease causing mutation, because 1) it was found in compound heterozygosity with p.R1939Q, 2) it was absent in controls, 3) it affects a C2 domain, and 4) the scores provided by prediction programs also agree with the pathogenicity. The pathogenic potential of the three other variants (p.D450E, p.V1778I, and p.E1803A) is less clear, because 1) all of them have been found in the heterozygous state without accompanying mutation in the other allele, and 2) p.D450E was found in controls. But it is also true that 1) they affect C2 domains, and 2) the scores of the prediction programs would support their classification as pathogenic variants.
As with other genes, the spectrum of OTOF mutations found in the Japanese population was quite different from those reported in Caucasians [13, 26–28].
With regard to recurrent mutations, p.Q829X especially has a high frequency in Spanish people, being present in about 3% of all cases of recessive prelingual deafness [24]. C.2905-2923delinsCTCCGAGCGGCA is also common in Argentineans [12] and p.E1700Q is reported to be frequent in Taiwanese [29]. p.R1939Q, previously identified in the United States [22] and most recently reported as a frequent mutation in Japanese [13], was also frequently found in this study. Among 160 patients, 8 (5.0%) had this mutation, confirming it is indeed a recurrent mutation in Japanese.
Those recurrent mutations have been proved to be due to founder effects [13, 24, 29].
Out of 16 patients with OTOF mutations, 7 showed ANSD phenotype, confirming that OTOF mutations are major causes of ANSD. In this study, 9 were heterozygous without second mutation. A hallmark of recessive mutations is the detection of two mutations in the paternal and maternal alleles and the parents having normal hearing. As seen in previous mutation screening reports, including those for OTOF[12, 23, 30], there were a significant number of heterozygous cases without a second mutation even after direct sequencing of the coding region of the gene. Possible explanations are: 1) the existence of a second mutation in the intron or regulatory region of OTOF, which has not been explored, 2) the existence of a large deletion [31], 3) contribution to hearing loss by an additional modulatory gene, and 4) the existence of a mutation in another gene and just coincidental carrying of the OTOF mutation.
As seen in Table 3, two heterozygous patients (#8, 9) having the ANSD phenotype, are most likely to have OTOF related deafness.
It is assumed that OTOF mutations accounted for deafness in at least 7, and possibly 16, of the 160 patients (4.4-10.0%). As described in the subject section, we excluded the subjects carrying GJB2 and SLC26A4 mutations. We also excluded another responsible gene (PJVK), because no mutations in this gene were found. Since the frequencies of GJB2 and SLC26A4 gene mutations among the patients with nonsyndromic severe to profound congenital SNHL are 27.0% based on our database, mutation frequency of OTOF among the total of severe to profound recessive nonsyndromic SNHL is considered to be about 3.2-7.3% (which is calculated by ((7-16)/160×(100/73))×100%). Although simple comparison regarding frequency is difficult because of sampling bias, it is estimated that the frequency of OTOF mutations in Japanese may be almost equal to other populations, as mutation frequency of OTOF was reported at 2.3% (13/557) in Pakistanis [11], 5.0% in Turkish [32], 1.4% (1/73) in Chinese [23], and 18.2% (4/22) in Taiwanese [29], and 3.2% (23/708) in Spanish [12]. Although simple comparison regarding frequency is difficult because of sampling bias, it is estimated that the frequency of OTOF mutations in Japanese may be almost equal to other populations. In Japanese, GJB2, SLC26A4, CDH23 and the 1555A>G mutation in the mitochondrial 12S rRNA are the major causes of hearing loss [33]. Considering the frequency, the OTOF gene may be one of the candidate genes to be screened for recessive severe to profound recessive SNHL.
The benefits of cochlear implantation for patients with ANSD has varied [34, 35], but implantation has been shown to be effective for the patients with OTOF mutations [15, 16, 36], because their auditory nerves and spiral ganglions are preserved. Consequently, if an OTOF mutation is identified in a deaf patient, we can anticipate a good outcome of cochlear implantation, therefore, it is important and meaningful to identify genetic mutations in patients.
Most patients with OTOF mutations have a phenotype of stable prelingual and severe to profound nonsyndromic hearing loss. On the other hand, other phenotypes have also been reported. For example, a Taiwanese patient with an p.E1700Q mutation displayed moderate to profound progressive hearing loss [29]. Temperature sensitive ANSD, a particular form of ANSD, has also been reported in some populations [10, 23, 37].
In the very young child, electrophysiological testing may indicate that OTOF-related deafness is ANSD, but by age two OAEs have generally disappeared and the test results are more in accord with the findings of cochlear lesions [14]. Therefore, if OAE is not tested at a very early age, patients with OTOF mutations are not deemed to have ANSD (i.e., hidden ANSD). In fact, 9 out of our 16 patients were diagnosed genetically as nonsyndromic sensorineural hearing loss (NSHL). According to the present data, screening for OTOF is necessary not only for the patients diagnosed with ANSD, but also should be extended to ARNSHL cases. The current data indicated that OAE testing must always be conducted in addition to ABR in infants. And we should bear in mind that there may be patients with OTOF mutations among the patients diagnosed as having ARNSHL.
Conclusions
The present study showed that OTOF mutations accounted for 3.2-7.3% of recessive severe to profound SNHL patients in Japan. OTOF mutations are a frequent cause in the Japanese deafness population and mutation screening should be considered regardless of the presence/absence of OAEs.
References
Starr A, Picton TW, Sininger Y, Hood LJ, Berlin CI: Auditory neuropathy. Brain. 1996, 119 (Pt 3): 741-753.
Kaga K, Nakamura M, Shinogami M, Tsuzuku T, Yamada K, Shindo M: Auditory nerve disease of both ears revealed by auditory brainstem responses, electrocochleography and otoacoustic emissions. Scand Audiol. 1996, 25 (4): 233-238. 10.3109/01050399609074960.
Berlin CI, Hood L, Morlet T, Rose K, Brashears S: Auditory neuropathy/dys-synchrony: diagnosis and management. Ment Retard Dev Disabil Res Rev. 2003, 9 (4): 225-231. 10.1002/mrdd.10084.
Roush P, Frymark T, Venediktov R, Wang B: Audiological Management of Auditory Neuropathy Spectrum Disorder in Children: A Systematic Review of the Literature. Am J Audiol. 2011, 159-170. 20
Madden C, Rutter M, Hilbert L, Greinwald JH, Choo DI: Clinical and audiological features in auditory neuropathy. Arch Otolaryngol Head Neck Surg. 2002, 128 (9): 1026-1030. 10.1001/archotol.128.9.1026.
Starr A, Sininger YS, Pratt H: The varieties of auditory neuropathy. J Basic Clin Physiol Pharmacol. 2000, 11 (3): 215-230.
Manchaiah VK, Zhao F, Danesh AA, Duprey R: The genetic basis of auditory neuropathy spectrum disorder (ANSD). Int J Pediatr Otorhinolaryngol. 2011, 75 (2): 151-158. 10.1016/j.ijporl.2010.11.023.
Roux I, Safieddine S, Nouvian R, Grati M, Simmler MC, Bahloul A, Perfettini I, Le Gall M, Rostaing P, Hamard G, et al: Otoferlin, defective in a human deafness form, is essential for exocytosis at the auditory ribbon synapse. Cell. 2006, 127 (2): 277-289. 10.1016/j.cell.2006.08.040.
Yasunaga S, Grati M, Chardenoux S, Smith TN, Friedman TB, Lalwani AK, Wilcox ER, Petit C: OTOF encodes multiple long and short isoforms: genetic evidence that the long ones underlie recessive deafness DFNB9. Am J Hum Genet. 2000, 67 (3): 591-600. 10.1086/303049.
Varga R, Avenarius MR, Kelley PM, Keats BJ, Berlin CI, Hood LJ, Morlet TG, Brashears SM, Starr A, Cohn ES, et al: OTOF mutations revealed by genetic analysis of hearing loss families including a potential temperature sensitive auditory neuropathy allele. J Med Genet. 2006, 43 (7): 576-581.
Choi BY, Ahmed ZM, Riazuddin S, Bhinder MA, Shahzad M, Husnain T, Griffith AJ, Friedman TB: Identities and frequencies of mutations of the otoferlin gene (OTOF) causing DFNB9 deafness in Pakistan. Clin Genet. 2009, 75 (3): 237-243. 10.1111/j.1399-0004.2008.01128.x.
Rodriguez-Ballesteros M, Reynoso R, Olarte M, Villamar M, Morera C, Santarelli R, Arslan E, Meda C, Curet C, Volter C, et al: A multicenter study on the prevalence and spectrum of mutations in the otoferlin gene (OTOF) in subjects with nonsyndromic hearing impairment and auditory neuropathy. Hum Mutat. 2008, 29 (6): 823-831. 10.1002/humu.20708.
Matsunaga T, Mutai H, Kunishima S, Namba K, Morimoto N, Shinjo Y, Arimoto Y, Kataoka Y, Shintani T, Morita N, et al: A prevalent founder mutation and genotype-phenotype correlations of OTOF in Japanese patients with auditory neuropathy. Clin Genet. 2012, 82 (5): 425-432. 10.1111/j.1399-0004.2012.01897.x.
Smith RJH, Gurrola JG, Kelley PM: OTOF-Related Deafness. GeneReviews [internet]. Edited by: Pagon RA, Adam MP, Bird TD, Dolan CR, Fong CT, Stephens K. 2008, Seattle, WA: University of Washington, updated 2011
Rouillon I, Marcolla A, Roux I, Marlin S, Feldmann D, Couderc R, Jonard L, Petit C, Denoyelle F, Garabedian EN, et al: Results of cochlear implantation in two children with mutations in the OTOF gene. Int J Pediatr Otorhinolaryngol. 2006, 70 (4): 689-696. 10.1016/j.ijporl.2005.09.006.
Wu CC, Liu TC, Wang SH, Hsu CJ, Wu CM: Genetic characteristics in children with cochlear implants and the corresponding auditory performance. Laryngoscope. 2011, 121 (6): 1287-1293. 10.1002/lary.21751.
Hilgert N, Smith RJ, Van Camp G: Forty-six genes causing nonsyndromic hearing impairment: which ones should be analyzed in DNA diagnostics?. Mutat Res. 2009, 681 (2–3): 189-196.
Wang K, Li M, Hakonarson H: ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38 (16): e164-10.1093/nar/gkq603.
Liu X, Jian X, Boerwinkle E: dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat. 2011, 32 (8): 894-899. 10.1002/humu.21517.
Chang X, Wang K: wANNOVAR: annotating genetic variants for personal genomes via the web. J Med Genet. 2012, 49 (7): 433-436. 10.1136/jmedgenet-2012-100918.
Wang J, Fan YY, Wang SJ, Liang PF, Wang JL, Qiu JH: Variants of OTOF and PJVK genes in Chinese patients with auditory neuropathy spectrum disorder. PLoS One. 2011, 6 (9): e24000-10.1371/journal.pone.0024000.
Varga R, Kelley PM, Keats BJ, Starr A, Leal SM, Cohn E, Kimberling WJ: Non-syndromic recessive auditory neuropathy is the result of mutations in the otoferlin (OTOF) gene. J Med Genet. 2003, 40 (1): 45-50. 10.1136/jmg.40.1.45.
Wang DY, Wang YC, Weil D, Zhao YL, Rao SQ, Zong L, Ji YB, Liu Q, Li JQ, Yang HM, et al: Screening mutations of OTOF gene in Chinese patients with auditory neuropathy, including a familial case of temperature-sensitive auditory neuropathy. BMC Med Genet. 2010, 11: 79-
Migliosi V, Modamio-Hoybjor S, Moreno-Pelayo MA, Rodriguez-Ballesteros M, Villamar M, Telleria D, Menendez I, Moreno F, Del Castillo I: Q829X, a novel mutation in the gene encoding otoferlin (OTOF), is frequently found in Spanish patients with prelingual non-syndromic hearing loss. J Med Genet. 2002, 39 (7): 502-506. 10.1136/jmg.39.7.502.
Mahdieh N, Shirkavand A, Rabbani B, Tekin M, Akbari B, Akbari MT, Zeinali S: Screening of OTOF mutations in Iran: a novel mutation and review. Int J Pediatr Otorhinolaryngol. 2012, 76 (11): 1610-1615. 10.1016/j.ijporl.2012.07.030.
Ohtsuka A, Yuge I, Kimura S, Namba A, Abe S, Van Laer L, Van Camp G, Usami S: GJB2 deafness gene shows a specific spectrum of mutations in Japan, including a frequent founder mutation. Hum Genet. 2003, 112 (4): 329-333.
Tsukamoto K, Suzuki H, Harada D, Namba A, Abe S, Usami S: Distribution and frequencies of PDS (SLC26A4) mutations in Pendred syndrome and nonsyndromic hearing loss associated with enlarged vestibular aqueduct: a unique spectrum of mutations in Japanese. Eur J Hum Genet. 2003, 11 (12): 916-922. 10.1038/sj.ejhg.5201073.
Wagatsuma M, Kitoh R, Suzuki H, Fukuoka H, Takumi Y, Usami S: Distribution and frequencies of CDH23 mutations in Japanese patients with non-syndromic hearing loss. Clin Genet. 2007, 72 (4): 339-344. 10.1111/j.1399-0004.2007.00833.x.
Chiu YH, Wu CC, Lu YC, Chen PJ, Lee WY, Liu AY, Hsu CJ: Mutations in the OTOF gene in Taiwanese patients with auditory neuropathy. Audiol Neurootol. 2010, 15 (6): 364-374. 10.1159/000293992.
Romanos J, Kimura L, Favero ML, Izarra FA, de Mello Auricchio MT, Batissoco AC, Lezirovitz K, Abreu-Silva RS, Mingroni-Netto RC: Novel OTOF mutations in Brazilian patients with auditory neuropathy. J Hum Genet. 2009, 54 (7): 382-385. 10.1038/jhg.2009.45.
Zadro C, Ciorba A, Fabris A, Morgutti M, Trevisi P, Gasparini P, Martini A: Five new OTOF gene mutations and auditory neuropathy. Int J Pediatr Otorhinolaryngol. 2010, 74 (5): 494-498. 10.1016/j.ijporl.2010.02.004.
Duman D, Sirmaci A, Cengiz FB, Ozdag H, Tekin M: Screening of 38 genes identifies mutations in 62% of families with nonsyndromic deafness in Turkey. Genet Test Mol Biomarkers. 2011, 15 (1–2): 29-33.
Usami SI, Nishio SY, Nagano M, Abe S, Yamaguchi T: Simultaneous screening of multiple mutations by invader assay improves molecular diagnosis of hereditary hearing loss: a multicenter study. PLoS One. 2012, 7 (2): e31276-10.1371/journal.pone.0031276.
Gibson WP, Sanli H: Auditory neuropathy: an update. Ear Hear. 2007, 28 (2 Suppl): 102S-106S.
Rance G, Barker EJ: Speech perception in children with auditory neuropathy/dyssynchrony managed with either hearing AIDS or cochlear implants. Otol Neurotol. 2008, 29 (2): 179-182. 10.1097/mao.0b013e31815e92fd.
Rodriguez-Ballesteros M, del Castillo FJ, Martin Y, Moreno-Pelayo MA, Morera C, Prieto F, Marco J, Morant A, Gallo-Teran J, Morales-Angulo C, et al: Auditory neuropathy in patients carrying mutations in the otoferlin gene (OTOF). Hum Mutat. 2003, 22 (6): 451-456. 10.1002/humu.10274.
Starr A, Sininger Y, Winter M, Derebery MJ, Oba S, Michalewski HJ: Transient deafness due to temperature-sensitive auditory neuropathy. Ear Hear. 1998, 19 (3): 169-179. 10.1097/00003446-199806000-00001.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2350/14/95/prepub
Acknowledgements
We thank all the families that participated in the present study. We would also like to thank Ms. S. Matsuda for technical assistance, Ms. A. C. Apple-Mathews for help in preparing the manuscript, and Dr. T. Matsunaga for valuable comments. This study was supported by a Health Sciences Research Grant from the Ministry of Health and Welfare of Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
YI and SN carried out the molecular genetic studies and the sequence alignment, and participated in drafting the manuscript. SU conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Iwasa, Yi., Nishio, Sy., Yoshimura, H. et al. OTOF mutation screening in Japanese severe to profound recessive hearing loss patients. BMC Med Genet 14, 95 (2013). https://doi.org/10.1186/1471-2350-14-95
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2350-14-95