Abstract
Background
HLA-DRB1 allele polymorphisms have been reported to be associated with hepatocellular carcinoma susceptibility, but the results of these previous studies have been inconsistent. The purpose of the present study was to explore whether specific HLA-DRB1 alleles (DRB1*07, DRB1*12, DRB1*15) confer susceptibility to hepatocellular carcinoma.
Methods
Case-control studies on HLA-DRB1 alleles association with HCC were searched up to January 2010 through a systematic review of the literature. The odds ratios (ORs) of HLA-DRB1 allele distributions in patients with hepatocellular carcinoma were analyzed against healthy controls. The meta-analysis software REVMAN 5.0 was applied for investigating heterogeneity among individual studies and for summarizing all the studies. Meta-analysis was performed using fixed-effect or random-effect methods, depending on absence or presence of significant heterogeneity.
Results
Eight case-control studies were included in the final analysis. Among the 3 HLA-DRB1 alleles studied, DRB1*07 and DRB1*12 were significantly associated with the risk of HCC in the whole populations (OR = 1.65, 95% CI: 1.08-2.51, P = 0.02 and OR = 1.59, 95% CI: 1.09-2.32, P = 0.02, respectively). No significant association was established for DRB1*15 allele with HCC in the whole populations. Subgroup analysis by ethnicity showed that DRB1*07, DRB1*12 and DRB1*15 alleles significantly increased the risk of hepatocellular carcinoma in Asians (OR = 2.10, 95% CI: 1.06-4.14, P = 0.03; OR = 1.73, 95% CI: 1.17-2.57, P = 0.006 and OR= 2.88, 95%CI: 1 .77-4.69, P < 0.001 , respectively).
Conclusion
These results support the hypothesis that specific HLA-DRB1 alleles might influence the susceptibility of hepatocellular carcinoma. Large, multi-ethnic confirmatory and well designed studies are needed to determine the host genetic determinants of hepatocellular carcinoma.
Similar content being viewed by others
Background
Hepatocellular carcinoma (HCC) is linked to the interaction between genetic, immunologic, environmental, dietary, and life style factors. Its incidence and distribution vary widely among ethnic groups, sex, and geographic regions. Hepatocellular carcinoma is the third most common cause of cancer-related deaths worldwide with about 600,000 patients dying from the disease annually [1]. Asian countries account for nearly 78% of the roughly 600,000 cases of hepatocellular carcinoma (HCC) reported globally each year [2]. China alone accounts for more than 50% of the world's cases [3]. HBV and HCV infection, liver cirrhosis, male gender, and old age are important risk factors of HCC. The clustering of HCC within families raises the possibility that genetic factors are also involved in susceptibility to HCC.
The Major Histocompatibility Complex (MHC) plays a key role in anti-virus and tumor defense. Human leukocyte antigens (HLA) function in the regulation of immune response to foreign antigens and discrimination of self from non-self antigens. They are encoded by a series of closely linked genetic loci found on chromosome 6 [4, 5]. HLA polymorphism is implicated in conferring genetic susceptibility to a large number of immune mediated diseases, including some cancers. Given the pivotal role of HLA molecules in the immune system, several studies have been performed to investigate the association between specific HLA alleles and HCC. However, the association between HLA-DRB1 alleles and HCC in different ethnic populations that has been reported is controversial. Many conflicting reports have been published to date; thus, we performed a systematic review of all of the relevant studies published in the literature to evaluate the association between HLA-DRB1 alleles and HCC. Our principal objectives were to clarify the specific HLA-DRB1 alleles that conferred susceptibility to or which protected against HCC.
Methods
Search strategy
Electronic databases (PubMed, EMBASE, Cochrane Library and China National Knowledge Infrastructure) were used to search for all genetic association studies evaluating the HLA-DRB1 polymorphism and HCC in humans in all languages up to January 2010. The search strategy was based on combinations of the terms: HLA-DRB1 AND (Hepatocellular carcinoma or HCC) AND (variants or polymorphism or alleles). We also performed a full manual search from the bibliographies of selected papers. We also contacted the authors of studies containing relevant information, who did not report the results necessary for this analysis. Unpublished data were also accepted if an abstract was available and further information was obtained from the author.
Selection criteria
In the meta-analysis, the following inclusive selection criteria were set and reviewed by two independent investigators: (1) each trial is an independent case-control study; (2) the purpose of all studies and statistical methods is similar; (3) it supplied enough information to calculate the odds ratio (OR);(4) HLA-DRB1 alleles were molecularly typed (high or low resolution level); (5) inclusion of patients according to the diagnosis standard of HCC defined in 2002, based on at least one of the following criteria: classical histological characteristics or serum a-fetoprotein (AFP) level higher than 400 ng/ml together with radiological findings (ultrasound and/or CT) consistent with HCC [6]. A single study, Donaldson et al, done before 2002, was included in the meta-analysis given that the inclusion criteria of patients were similar to the diagnosis standard. The following exclusive selection criteria were set: (1) incomplete raw data; (2) repetitive reports (if more than one version of the same study was retrieved, only the most recent is used); (3) materials and methods were not well-described and reliable.
Although assessment of study quality is considered important for systematic reviews and meta-analyses, scoring methods have been considered problematic [7] and may not accurately assess the quality measures of interest [8]. Therefore, we used reliability of patient selection, molecular typing method, and statistical analysis method as quality variables.
The frequency of HLA-DRB1 alleles varies according to ethnic and racial background, with some alleles being extremely rare. Therefore, articles were not required to identify all alleles for inclusion.
Data extraction
The studies were independently evaluated by two researchers. Discrepancies in the evaluations of some studies were resolved by discussion between the reviewers. The following data were collected from each study: authors, publication year, journal, publication type and language, HLA genotyping method, allele genotyped, allele frequencies, numbers of cases and controls, definitions criteria used for HCC, HCC sample description, controls sample description. Allelic frequency was calculated as the number of cases or controls harboring at least one allele type (HLA-DRB1) divided by the total number of chromosomes included in each of the corresponding groups.
Statistical analysis
The literature review conformed to PRISMA statement standards, and our research fit the minimum set of items for reporting in systematic reviews and meta-analyses (Additional file 1). Heterogeneity was calculated by means of Cochran's Q test (α = 0.05) and Higgins's (I 2 ) tests. I 2 values of 25%, 50% and 75% were assigned as low, moderate, and high estimates, respectively. If the results of the Q test had no significant heterogeneity, the Mantel-Haenszel fixed effect model (Peto method) was used for the combination of data; If the results of the Q test had significant heterogeneity, the Dersimonian-Laird random effects model (DL method) were used for the combination of data [9]. A pooled OR was presented as a standard plot with 95% confidence intervals (95%CIs). Meta-analysis was performed using fixed-effect or random-effect methods, depending on absence or presence of significant heterogeneity. As a measure of association between HCC and HLA-DRB1 alleles, we combined odds ratios (ORs) with 95% confidence intervals (95%CIs) stratified by gene subtype of patients and controls in a study. Funnel plots and the Egger's regression asymmetry test were used to evaluate publication bias [10]. All P values presented are two-tailed. To reduce heterogeneity and to evaluate whether there was a different genotype effect in predefined subgroups of studies, we performed subgroup analysis according to the ethnicity. We performed a sensitivity analysis to assess the stability of the results by sequential omission of individual studies. The analyses were performed using Revman 5.0 provided by the Cochrane Collaboration.
Results
Literature assessment
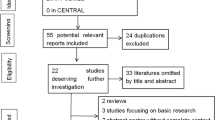
Figure 1 shows the flow chart of publications identified by the literature search. Search strategy allowed us to identify 84 studies for potential inclusion in the meta-analysis. Finally 8 case-control studies relating to HLA-DRB1 alleles polymorphism and susceptibility to HCC qualified on the basis of our selection criteria [11–18]. A total of 957 subjects were studied (including 341 patients and 616 controls). The main features of the studies included in the meta-analysis are shown in Table 1. Among the eight studies, five studies were conduct in Asian countries; two were conduct in European countries, and one was conduct in African countries. Mean or median age was not stated in 4/8 reports and sex in 4/8. HIV status was determined in only one report [12].
HLA-DRB1 alleles were molecularly typed (high or low resolution level). Five studies used low resolution molecular typing for HLA, while three used high resolution molecular typing for HLA. Low resolution molecular typing methods for HLA could not identify the specific alleles. Accurate methods for HLA class II typing should involve the combination of PCR-SSOP, PCR-SSP, and PCR-SSCP [19].
Meta-analysis: Association between HLA-DRB1 alleles with HCC
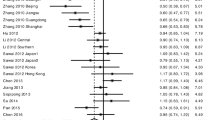
A total of 13 HLA-DRB1 alleles were studied in the 8 case-control studies, but only 3 alleles (DRB1*07, DRB1*12, DRB1*15) was extracted from the studies to investigate their association with HCC, which were reported at least six of the eight case-control studies. Statistics calculated for each study are shown in the forest plot (Figures 2, 3 and 4).
In the meta-analysis, overall the frequencies of HLA-DRB1*07 allele was 20.1% (59 of 281) in HCC and 15.2% (71 of 466) in controls. The heterogeneity test indicates that the variation of trial-specific ORs was not statistically significant (χ2 = 10.79, I 2 = 54%, P = 0.06 and >0.05). Under the fixed effect model, the combined OR for the association of HLA-DRB1*07 allele with the risk for HCC in the whole populations was determined to be 1.65 (95% CI: 1.08-2.51; p = 0.02), and under the random effects model was 1.77 (95%CI: 0.88-3.56; p = 0.11). In sensitivity analysis, the exclusion of individual studies did not change this significant result, except for exclusion of the study by Pan et al and El-Chennawi et al, which produced a non-significant association. Subgroup analysis by ethnicity showed that HLA-DRB1*07 allele significantly increased the risk of hepatocellular carcinoma in Asians under the fixed effect model (OR = 2.10, 95% CI: 1.06-4.14, P = 0.03).
Overall the frequencies of HLA-DRB1*12 allele was 26.3% (74 of 281) in HCC and 16.1% (83 of 516) in controls. The heterogeneity test indicates that the variation of trial-specific ORs was not statistically significant (χ2 = 9.00, I 2 = 44%, P = 0.11 and >0.05), so the fixed-effect method was used to combine the results. The combined OR for the association of HLA-DRB1*12 allele with the risk for HCC in the whole populations was determined to be 1.59 (95% CI: 1.09-2.32), and was statistically significant (P = 0.02 and <0.05). In sensitivity analysis, the exclusion of individual studies did not change this significant result, except for exclusion of the study by Donaldson, et al and Sun, et al, which produced a non-significant association. Subgroup analysis by ethnicity showed that HLA-DRB1*12 allele significantly increased the risk of hepatocellular carcinoma in Asians (OR = 1.73, 95% CI: 1.17-2.57, P = 0.006).
Meta-analysis for HLA-DRB1*15 allele was carried out, but it did not show any statistical effect in the whole populations. Subgroup analysis by ethnicity showed that HLA-DRB1*15 allele significantly increased the risk of hepatocellular carcinoma in Asians under the fixed effect model (OR= 2.88, 95% CI: 1 .77-4.69, P < 0.001 ).
Figures 5, 6 and 7 show the funnel plot analysis to detect publication bias of each study for DRB1*07, DRB1*12 and DRB1*15, respectively. The shape of the funnel plot seemed to be asymmetrical, suggesting that publication bias might affect the findings of our meta-analysis.
Discussion
The aim of the present study was to ascertain whether specific HLA-DRB1 alleles are associated with the development/protection of HCC. We analyzed the published studies investigating the association between HLA-DRB1 alleles and HCC. Studies concerning this possible association have been undertaken since the early 1996s [20].
To our knowledge, this is the first published meta-analysis investigating this association. Our meta-analysis of eight studies revealed that DRB1*07 and DRB1*12 were significantly associated with the risk of HCC in the whole populations (OR = 1.65, 95% CI: 1.08-2.51, P = 0.02 and OR = 1.59, 95% CI: 1.09-2.32, P = 0.02, respectively). Subgroup analysis by ethnicity showed that DRB1*07 and DRB1*12 alleles also significantly increased the risk of hepatocellular carcinoma in Asians (OR = 2.10, 95% CI: 1.06-4.14, P = 0.03; OR = 1.73, 95% CI: 1.17-2.57, P = 0.006, respectively). No significant association was established for DRB1*15 allele with HCC in the whole populations. Subgroup analysis by ethnicity showed that DRB1*15 alleles significantly increased the risk of hepatocellular carcinoma in Asians under the fixed effect model (OR= 2.88, 95% CI: 1 .77-4.69, P < 0.001 ).
Epidemiological survey showed that Asian countries account for nearly 78% of hepatocellular carcinoma (HCC) reported globally each year, and Hepatitis B Virus (HBV) is the major etiology of HCC in these areas. Although HBV infection plays an important role in HCC, HBV infection alone is not sufficient for progression to HCC. Several lines of evidence suggest that cellular immune surveillance is important in the control of HBV infection and the development of HCC. In 2007, Yang and his colleagues found that HLA-DRB1*07 were markedly higher in the HBV-infected group among people in northwestern China (17.6% of HBV-infected patients vs 9.3% of spontaneously cleared controls, OR = 2.09, P < 0.05) [21]. In 2006, Zhang and his colleagues found that the frequency of HLA-DRB1*12 was significantly higher in the HBV persistent group than in the recovered group among Chinese (0.230 versus 0.063, P = 0.004, OR = 2.09) [22]. In 2003, Amarapurpar and his colleagues found that a positive association of HLA-DRB1*15 to persistence of HBV among Indians (57.6 vs. 25%) [23]. As we know that clearance of acute hepatitis B virus (HBV) infection is associated with a vigorous CD4+ T-cell response focusing on the core protein. HLA class II glycoproteins present viral peptides to CD4+ T cells and influence the immune responses. Binding affinities of overlapping peptides covering the core and envelope proteins of HBV were measured to HLA glycoproteins encoded by some HLA-DRB1 molecules and compared with published peptide-specific CD4+ T-cell responses [24]. So we have a hypothesis that HLA-DRB1*07, DRB1*12 and DRB1*15 alleles may be the key host factors to determine the development of diseases from HBV infection to HCC in Asians, basing on our results that HLA-DRB1*07, DRB1*12 and DRB1*15 alleles significantly increased the risk of hepatocellular carcinoma in Asians. Furthermore, the importance of environmental factors and gene-environmental interactions in the development of HCC should not be ignored and is beginning to be delineated.
Moreover, HLA-DRB1 alleles polymorphism have been reported to be associated with other cancers and autoimmune diseases, including cervical squamous cell carcinoma, rheumatoid arthritis, systemic lupus erythematosus, autoimmune hepatitis, inflammatory bowel disease, multiple sclerosis and type 1 diabetes, and meta-analyses have been done for these diseases [25–31].
Additionally, because the information used in our research was based on data from observational studies, some limitations should be discussed in this meta-analysis and the results should be considered with caution. A primary cause for the difference in results by different authors may be related to the great variability of the frequency of HLA alleles in different populations. It is quite possible that one ethnic group may have some specific alleles in development/protection of HCC compared to other ethnic groups. Many studies were conducted on relatively smaller samples. Insufficient number of individuals might decrease the power to detect a difference in the distribution of DRB1*07, DRB1*12 and DRB1*15 alleles between HCC patients and controls, though a true difference exist. Lack of an association may not mean that associations do not exist. Many studies did not control for the matching variables in the analysis, and the possible confounders could be among the potential causes of variation in the studies' estimates. Different types of control groups could also be among the potential causes of variation in the studies' estimates. Effects of interactions in other environmental/behavioral and/or viral factors may be inevitable. A complex interplay between various genes is likely to modulate the development of HCC rather than a single allele. HLA genotyping techniques must be taken into consideration because these methodologies have different sensitivities. HLA genotyping were mainly tested for by PCR with probe hybridization, but targets were not always verified by sequencing. This could hinder an effective comparison between the studies and influence the combined results. The shape of the funnel plot seemed to be asymmetrical, suggesting that publication bias might affect the findings of our meta-analysis. Furthermore, although we tried to maximize our efforts to identify all relevant published studies in peer-reviewed journals, it is possible that some escaped our attention.
In spite of these, our meta-analysis also had some advantages. First, substantial number of cases and controls were pooled from different studies, which significantly increased statistical power of the analysis. Second, the quality of case-control studies included in the current meta-analysis was satisfactory based on our selection criteria. Third, the patients were phenotypic homogeneous subjects, diagnosed either by gold standard or by biochemical and imageology combined methods, which could reduce the heterogeneity in some extent.
Conclusion
Our meta-analysis suggests that HLA-DRB1*07 and DRB1*12 alleles are risk factors for HCC in the whole populations, especially in the Asians; DRB1*15 allele is only associated with an increased risk of HCC in Asians (under the fixed effect model). However, it is necessary to conduct large trials using standardized unbiased methods, homogeneous HCC patients and well matched controls, with the assessors blinded to the data. Moreover, gene-gene and gene-environment interactions should also be considered in the analysis. Such studies taking these factors into account may eventually lead to a better, more comprehensive understanding of the association between HLA-DRB1 polymorphism and HCC.
Abbreviations
- HLA:
-
human leukocyte antigens
- HCC:
-
hepatocellular carcinoma
- HBV:
-
hepatitis B virus
- OR:
-
odds ratio
- CI:
-
confidence interval.
References
Schutte K, Bornschein J, Malfertheiner P: Hepatocellular carcinoma--epidemiological trends and risk factors. Dig Dis. 2009, 27: 80-92.
International Agency for Cancer Research: GLOBCAN 2002. [http://www-dep.iarc.fr]
El-Serag HB, Rudolph KL: Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007, 132: 2557-2576. 10.1053/j.gastro.2007.04.061.
Klein J, Sato A: The HLA system. First of two parts. N Engl J Med. 2000, 343: 702-709. 10.1056/NEJM200009073431006.
Klein J, Sato A: The HLA system. Second of two parts. N Engl J Med. 2000, 343: 782-786. 10.1056/NEJM200009143431106.
Befeler AS, Di Bisceglie AM: Hepatocellular carcinoma: diagnosis and treatment. Gastroenterology. 2002, 122: 1609-1619. 10.1053/gast.2002.33411.
Conn VS, Rantz MJ: Research methods: managing primary study quality in meta-analyses. Res Nurs Health. 2003, 26: 322-333. 10.1002/nur.10092.
Huwiler-Muntener K, Juni P, Junker C, Egger M: Quality of reporting of randomized trials as a measure of methodologic quality. JAMA. 2002, 287: 2801-2804. 10.1001/jama.287.21.2801.
DerSimonian R, Laird N: Meta-analysis in clinical trials. Control Clin Trials. 1986, 7: 177-188. 10.1016/0197-2456(86)90046-2.
Egger M, Davey Smith G, Schneider M, Minder C: Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997, 315: 629-634.
Donaldson PT, Ho S, Williams R, Johnson PJ: HLA class II alleles in Chinese patients with hepatocellular carcinoma. Liver. 2001, 21: 143-148. 10.1034/j.1600-0676.2001.021002143.x.
De Re V, Caggiari L, Talamini R, Crovatto M, De Vita S, Mazzaro C, Cannizzaro R, Dolcetti R, Boiocchi M: Hepatitis C virus-related hepatocellular carcinoma and B-cell lymphoma patients show a different profile of major histocompatibility complex class II alleles. Hum Immunol. 2004, 65: 1397-1404. 10.1016/j.humimm.2004.08.183.
Yuan JH, Sun CG: Relationship between every kind of chronic hepatitis B' prognosis and HLA-DRB1 allele in Chinese patients of shandong area. Chin J Clini Hepatol. 2004, 20: 236-237.
Lopez-Vazquez A, Rodrigo L, Mina-Blanco A, Martinez-Borra J, Fuentes D, Rodriguez M, Perez R, Gonzalez S, Lopez-Larrea C: Extended human leukocyte antigen haplotype EH18.1 influences progression to hepatocellular carcinoma in patients with hepatitis C virus infection. J Infect Dis. 2004, 189: 957-963. 10.1086/382189.
Yuan JH, Cui M, Sun CG, Yu HL: Association between HLA-DRB1*1201/*1501 and diferent stages of HBV infection. J Shandong University. 2005, 43: 428-431.
Kummee P, Tangkijvanich P, Poovorawan Y, Hirankarn N: Association of HLA-DRB1*13 and TNF-alpha gene polymorphisms with clearance of chronic hepatitis B infection and risk of hepatocellular carcinoma in Thai population. J Viral Hepat. 2007, 14: 841-848.
El-Chennawi FA, Auf FA, Metwally SS, Mosaad YM, El-Wahab MA, Tawhid ZE: HLA-class II alleles in Egyptian patients with hepatocellular carcinoma. Immunol Invest. 2008, 37: 661-674. 10.1080/08820130802111605.
Pan HF, Li DF, Sun TH, Wang XJ: Association of HLA-DRB1 gene polymorphism and expression of DR antigen with primary hepatocellular carcinoma. Chin J Hepatobiliary Surg. 2009, 15: 357-361.
Thio CL, Thomas DL, Carrington M: Chronic viral hepatitis and the human genome. Hepatology. 2000, 31: 819-827. 10.1053/he.2000.4316.
Aikawa T, Kojima M, Onishi H, Tamura R, Fukuda S, Suzuki T, Tsuda F, Okamoto H, Miyakawa Y, Mayumi M: HLA DRB1 and DQB1 alleles and haplotypes influencing the progression of hepatitis C. J Med Virol. 1996, 49: 274-278. 10.1002/(SICI)1096-9071(199608)49:4<274::AID-JMV3>3.0.CO;2-0.
Yang G, Liu J, Han S, Xie H, Du R, Yan Y, Xu D, Fan D: Association between hepatitis B virus infection and HLA-DRB1 genotyping in Shaanxi Han patients in northwestern China. Tissue Antigens. 2007, 69: 170-175. 10.1111/j.1399-0039.2006.00744.x.
Zhang SY, Gu HX, Li D, Yang SF, Zhong ZH, Li XK, Jin X: Association of human leukocyte antigen polymorphism with hepatitis B virus infection and genotypes. Jpn J Infect Dis. 2006, 59: 353-357.
Amarapurpar DN, Patel ND, Kankonkar SR: HLA class II genotyping in chronic hepatitis B infection. J Assoc Physicians India. 2003, 51: 779-781.
Godkin A, Davenport M, Hill AV: Molecular analysis of HLA class II associations with hepatitis B virus clearance and vaccine nonresponsiveness. Hepatology. 2005, 41: 1383-1390. 10.1002/hep.20716.
Yang YC, Chang TY, Lee YJ, Su TH, Dang CW, Wu CC, Liu HF, Chu CC, Lin M: HLA-DRB1 alleles and cervical squamous cell carcinoma: experimental study and meta-analysis. Hum Immunol. 2006, 67: 331-340. 10.1016/j.humimm.2006.03.017.
Delgado-Vega AM, Anaya JM: Meta-analysis of HLA-DRB1 polymorphism in Latin American patients with rheumatoid arthritis. Autoimmun Rev. 2007, 6: 402-408. 10.1016/j.autrev.2006.11.004.
Castano-Rodriguez N, Diaz-Gallo LM, Pineda-Tamayo R, Rojas-Villarraga A, Anaya JM: Meta-analysis of HLA-DRB1 and HLA-DQB1 polymorphisms in Latin American patients with systemic lupus erythematosus. Autoimmun Rev. 2008, 7: 322-330.
Duarte-Rey C, Pardo AL, Rodriguez-Velosa Y, Mantilla RD, Anaya JM, Rojas-Villarraga A: HLA class II association with autoimmune hepatitis in Latin America: a meta-analysis. Autoimmun Rev. 2009, 8: 325-331. 10.1016/j.autrev.2008.11.005.
Stokkers PC, Reitsma PH, Tytgat GN, van Deventer SJ: HLA-DR and -DQ phenotypes in inflammatory bowel disease: a meta-analysis. Gut. 1999, 45: 395-401. 10.1136/gut.45.3.395.
Rojas OL, Rojas-Villarraga A, Cruz-Tapias P, Sanchez JL, Suarez-Escudero JC, Patarroyo MA, Anaya JM: HLA class II polymorphism in Latin American patients with multiple sclerosis. Autoimmun Rev. 2010, 9: 407-413. 10.1016/j.autrev.2009.11.001.
Zhang XM, Wang HY, Luo YY, Ji LN: HLA-DQ, DR allele polymorphism of type 1 diabetes in the Chinese population: a meta-analysis. Chin Med J (Engl). 2009, 122: 980-986.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-230X/10/145/prepub
Acknowledgements
This work was partially supported by grants from the Natural Science Foundation of Shandong Province, China (No. ZR2009CQ031)
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
ZHL and YNX carried out the design of this meta-analysis, conducted a searching, extracted data, analyzed the data and drafted the manuscript. SYX participated in study design and the critical revision of the manuscript. QJD, QW, SHZ, XJJ and YS participated in the critical revision of the manuscript. All authors read and approved the final manuscript.
Zhong-Hua Lin, Yong-Ning Xin contributed equally to this work.
Electronic supplementary material
12876_2010_535_MOESM1_ESM.DOC
Additional file 1: PRISMA 2009 Checklist. We conformed to PRISMA statement standards and provided the detailed information. (DOC 69 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Lin, ZH., Xin, YN., Dong, QJ. et al. Association between HLA-DRB1 alleles polymorphism and hepatocellular carcinoma: a meta-analysis. BMC Gastroenterol 10, 145 (2010). https://doi.org/10.1186/1471-230X-10-145
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-230X-10-145