Abstract
Background
Enterococcus faecium has globally emerged as a cause of hospital-acquired infections with high colonization rates in hospitalized patients. The enterococcal surface protein Esp, identified as a potential virulence factor, is specifically linked to nosocomial clonal lineages that are genetically distinct from indigenous E. faecium strains. To investigate whether Esp facilitates bacterial adherence and intestinal colonization of E. faecium, we used human colorectal adenocarcinoma cells (Caco-2 cells) and an experimental colonization model in mice.
Results
No differences in adherence to Caco-2 cells were found between an Esp expressing strain of E. faecium (E1162) and its isogenic Esp-deficient mutant (E1162Δesp). Mice, kept under ceftriaxone treatment, were inoculated orally with either E1162, E1162Δesp or both strains simultaneously. Both E1162 and E1162Δesp were able to colonize the murine intestines with high and comparable numbers. No differences were found in the contents of cecum and colon. Both E1162 and E1162Δesp were able to translocate to the mesenteric lymph nodes.
Conclusion
These results suggest that Esp is not essential for Caco-2 cell adherence and intestinal colonization or translocation of E. faecium in mice.
Similar content being viewed by others
Background
Enterococci are normal inhabitants of the human gastrointestinal (GI) tract, but have emerged as important nosocomial pathogens with high-level resistance to antibiotics, such as ampicillin, aminoglycosides, and vancomycin [1]. They can cause a wide spectrum of diseases, including bacteremia, peritonitis, surgical wound infections, urinary tract infections, endocarditis, and a variety of device-related infections [1–11]. The majority of the enterococcal infections are caused by Enterococcus faecalis. However, in parallel with the increase in nosocomial enterococcal infections, a partial replacement of E. faecalis by Enterococcus faecium has occurred in European and United States hospitals [12–14]http://www.earss.rivm.nl.
Molecular epidemiological studies indicated that E. faecium isolates responsible for the majority of nosocomial infections and hospital outbreaks are genetically distinct from indigenous intestinal isolates [15, 16]. Recent studies revealed intestinal colonization rates with these hospital-acquired E. faecium as high as 40% in hospital wards, while colonization in healthy people appeared to be almost absent [13, 15, 16]. It is assumed that adherence to mucosal surfaces is a key process for bacteria to survive and colonize the GI tract. Intestinal colonization of nosocomial E. faecium strains is a first and key step that precedes clinical infection due to fecal contamination of catheters or wounds, and in the minority of infections, through bacterial translocation from the intestinal lumen to extraintestinal sites [17, 18]. It is not known which factors facilitate intestinal colonization of nosocomial E. faecium strains. The enterococcal surface protein Esp, located on a putative pathogenicity island [19, 20], is specifically enriched in hospital-acquired E. faecium and has been identified as a potential virulence gene. Esp is involved in biofilm formation [21] and its expression is affected by changes in environmental conditions, being highest in conditions that mimic the microenvironment of the human large intestines: 37°C and anaerobioses [22]. Furthermore, in one study, bloodstream isolates of E. faecium enriched with esp had increased adherence to human colorectal adenocarcinoma cells (Caco-2 cells) [23], suggesting a role of Esp in intestinal colonization. In contrast, adherence of E. faecium to Caco-2 cell lines was not associated with the presence of esp in another study [24]. In E. faecalis, Esp is also located on a pathogenicity island, although the genetic content and organization of the E. faecium and E. faecalis PAI is different. Esp of E. faecalis is also expressed on the surface of the bacterium [25, 26] and is important in colonization of urinary tract epithelial cells [25]. By using a mouse model, Pultz et al. [27] showed that Esp does not facilitate intestinal colonization or translocation of E. faecalis in mice, however this does not automatically predict a lack function for E. faecium Esp in murine colonization. First data suggest that the function of Esp in both enterococcal species might be different. Esp of E. faecium is clearly involved in biofilm formation (see above) while there is controversy about the role of E. faecalis Esp in biofilm formation [28–31]. Furthermore, studies so far indicate that E. faecalis harbors more virulence determinants then E. faecium. For instance, besides Esp different determinants (GelE, BopD, fsr locus, and bee locus) are putatively involved in biofilm formation [32–34]. This suggests that virulence factors in E. faecalis play somewhat redundant or partially overlapping roles such that the absence of a single virulence factor, like Esp, has only minimal effect. To elucidate the role of Esp of E. faecium in bacterial adhesion and intestinal colonization, we studied an Esp mutant, constructed and described recently [21], and its Esp expressing parent strain for their ability to adhere to intestinal epithelial cells and intestinal colonization by using Caco-2 cells and a mouse model.
Results
Adherence assay to Caco-2 cells
To determine whether Esp contributes to adherence of intestinal epithelial cells, the Esp expressing E. faecium strain E1162, its isogenic Esp-deficient mutant (E1162Δesp), and an E. faecium esp-negative strain (E135) were investigated for their ability to adhere to differentiated 14 days old Caco-2 cells. Strain E1162 exhibited high adherence to Caco-2 cells, while the esp-negative strain, E135, showed only low-level binding to Caco-2 cells (Figure 1). This difference in adherence was significant (P < 0.005). However, no significant difference in adherence to Caco-2 cells was observed between E1162 and E1162Δesp.
Intestinal colonization
To investigate the role of Esp in intestinal colonization and translocation to MLN, the Esp expressing E1162 and its isogenic Esp-deficient mutant (E1162Δesp) were inoculated orally in mice separately or simultaneously in a mixed inoculum. Mice were kept under ceftriaxone treatment the entire experiment. Prior to any intervention no E. faecium was cultured from stools of mice. The mean enterococcal contents of the stool of naïve mice was 5 × 105 ± 2 × 105 CFU/gram, these colonies were specified being E. faecalis.
Both E1162 and E1162Δesp were able to colonize the intestinal tract with comparable high numbers of cells for the entire 10 days of the experiment. One day after inoculation E1162 reached a median of 5.2 (range 2–15) × 108 CFU/gram of stool and E1162Δesp of 5.1 (1.6 – 8.2) × 108 CFU/gram. Ten days after inoculation, the amount of both strains slightly reduced to 3.7 (1.3–10) × 106 and 2.7 (0.2–25) × 106 CFU/gram of stool, respectively (Figure 2A). Similar amounts of E1162 and E1162Δesp were found in the stool of mice colonized when the mixed inoculum was administered (data not shown). After 10 days of colonization, all mice were sacrificed and E. faecium colonies obtained from small bowel, cecum, and colon contents were calculated. In both cecum and colon comparable amounts of E1162 (cecum contents 6.9 (0.04–7.3) × 106 and colon contents 3.9 (1.3–11) × 106 CFU/gram) and E1162Δesp (cecum contents 10 (0.4–200) × 106 and colon contents 2.7 (0.2–24) × 106 CFU/gram) were isolated, from both separate (Figure 2B) and mixed inocula (data not shown). Significantly more E1162Δesp (8.4 (0.5–300) × 106 CFU/gram) compared to E1162 (6.5 (0.5–52) × 104 CFU/gram) was isolated from the small bowel contents of mice when inoculated separately with E1162 wild type and the Esp-mutant strain (p = 0.002). This difference was not found in mice inoculated with the mixture of E1162 and E1162Δesp (data not shown).
Intestinal colonization. Mice were orally inoculated with E1162 (black circles) or E1162Δesp (open circles). (A) Numbers of E1162 and E1162Δesp were determined in stool of mice at different time points after E. faecium inoculation. (B) After 10 days of colonization, numbers of E1162 and E1162Δesp were determined in small bowel, cecum and colon. Data are expressed as CFU per gram of stool/fecal contents and medians are shown for 7 mice per group.
Both E1162 and E1162Δesp were able to translocate to the MLN. From both of the separately inoculated groups of mice, three out of seven MLN were found positive for either E1162 or E1162Δesp. No bacteria were cultured from blood. No pathological changes in the intestinal wall were observed in any of the colonized mice.
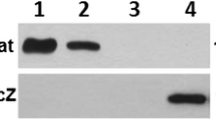
For both mono infection and mixed infection, randomly picked colonies were tested by MLVA to confirm strain identity. All colonies had the same MLVA profile belonging to E. faecium E1162(Δesp).
Discussion
Nosocomial E. faecium infections are primarily caused by specific hospital-selected clonal lineages, which are genetically distinct from the indigenous enterococcal flora. High rates of colonization of the GI tract of patients by these hospital-selected lineages upon hospitalization have been documented [13, 15]. Once established in the GI tract these nosocomial strains can cause infections through bacterial translocation from the GI tract to extraintestinal sites [35, 36]. The mechanism which promotes supplementation of the commensal enterococcal population by these nosocomial strains is not known. Destabilization of the GI tract through antibiotic therapy may provide nosocomial strains enhanced opportunities to gain a foothold in the GI tract. However, the effect of antibiotics is probably not the sole explanation for the emergence of nosocomial E. faecium infections since many antibiotics used in hospitals have relatively little enterococcal activity. This implicates that nosocomial E. faecium strains may possess traits that facilitate colonization of portions of the GI tract that the indigenous flora cannot effectively monopolize. Cell surface proteins like Esp, implicated in biofilm formation and specifically enriched in nosocomial strains, could represent one of these traits. Previously, it was shown that E. faecium is able to adhere to human and mouse intestinal mucus in vitro and becomes associated in vivo with the intestinal mucus layer of clindamycin treated mice [37–39]. This suggests an interaction between the bacterium and the mucus or with the epithelium itself. To examine the role of Esp in intestinal adherence and colonization, an Esp expressing strain of E. faecium (E1162) and its isogenic Esp-deficient mutant (E1162Δesp) were studied for adherence to differentiated Caco-2 cells and colonization of murine intestines. E1162, a hospital-acquired strain, exhibited significantly higher adherence to Caco-2 cells than E135, a representative of the indigenous flora. These results are consistent with an earlier study performed by Lund et al. [23]. However, no difference in adherence to Caco-2 cells between the E1162 and the E1162Δesp was found, indicating that Esp is not the determining factor responsible for the observed difference in Caco-2 cell adherence between nosocomial and indigenous E. faecium strains. This also implies that other determinants present in hospital-acquired E. faecium strains contribute to adhesion to intestinal epithelial cells. Comparative genomic hybridizations of 97 E. faecium nosocomial, commensal and animal isolates identified more than 100 genes that were enriched in nosocomial strains, including genes encoding putative adhesins, antibiotic resistance, IS elements, phage sequences, and novel metabolic pathways [40].
In addition, similar levels of intestinal colonization or translocation were found after inoculation with E1162 wild type or the isogenic Esp mutant E1162Δesp. These data are in accordance with a study performed by Pultz et al. [27] in which they showed that Esp did not facilitate intestinal colonization or translocation of E. faecalis in clindamycin-treated mice. Only from the small bowel contents of mice when inoculated separately with E1162 wild type and the Esp-mutant strain significantly more E1162Δesp compared to E1162 was isolated. This was an unexpected observation and we have no explanation for the fact that the levels of E1162Δesp in the small bowel are as high as in the cecum. Relatively lower levels as seen for E1162 are more typical for the small bowel.
Conclusion
Our data clearly demonstrate that Esp is not essential for high density colonization of the GI tract by nosocomial strains. Other possible candidate traits implicated in this process could include novel adhesins, like the novel cell surface proteins recently identified [41], bacteriocins, factors that resist specific or non-specific host defence mechanisms, and/or the ability to utilize new growth substrates. It is interesting in this respect that we recently identified a novel genomic island highly specific for nosocomial strains that tentatively encodes novel sugar uptake system [42].
For nosocomial E. faecium clones the GI tract serves as staging area from which they can disperse in and between patients, ultimately causing hospital-wide outbreaks. It is therefore of utmost importance to gain insight into the processes and determinants that promote intestinal colonization of nosocomial E. faecium strains. Only then we will be able to impede subsequent spread of these nosocomial clones.
Methods
Bacterial strains and growth conditions
In this study E. faecium strains E135, E1162 and E1162Δesp were used. E135 is an esp negative community surveillance feces isolate, while strain E1162 is a hospital-acquired blood isolate, positive for Esp expression. The isogenic Esp-deficient mutant, E1162Δesp, was previously constructed by introduction of a chloramphenicol resistance cassette (cat) resulting in an insertion-deletion mutation of the esp gene [21].E. faecium strains were grown in either Todd-Hewitt (TH) or Brain Heart Infusion (BHI) broth or on Tryptic Soy Agar (TSA) with 5% sheep red blood cells (Difco, Detroit, MI). Slanetz and Bartley (SB) agar plates were used to selectively grow enterococci. E. faecium strain E1162 and its isogenic mutant are high-level resistant to ceftriaxone (minimum inhibitory concentration > 32 μg/ml).
Caco-2 cell cultures
Human colorectal adenocarcinoma cells, Caco-2 cells, were obtained from the American Type Culture Collection (HTB-37, ATCC, USA) and were cultured in Dulbecco's Modified Eagle Medium (DMEM; Gibco, Invitrogen, Paisley, UK) supplemented with 10% heat-inactivated fetal calf serum (Integro B.V, Zaandam, The Netherlands), 1% non-essential amino acids (Gibco), 2 mM glutamine (Gibco), and 50 μg/ml gentamicin (Gibco). Cells were collected every 7th day by washing the monolayer twice with 0.022% disodium-ethylenediamine tetra acetic acid (di-Na-EDTA; Acros Organics, Morris Plains, NJ) in PBS and trypsinizing the cells using 50 μg/ml trypsine (Gibco), in 0.022% di-Na-EDTA in PBS. Cells were seeded at 1 × 106 cells in 10 ml DMEM in 75 cm2 culture bottles (Costar, Corning, NY) and incubated in a humidified, 37°C incubator with 5% CO2. The culture medium was refreshed every 4th day after passage of the cells. Differentiated Caco-2 cells were prepared by seeding cells from passage 25 to 45 in 12-wells tissue culture plates (Costar) at 1.6 × 105 cells/ml in DMEM. To each well 1 ml of this suspension was added and plates were incubated at 37°C with 5% CO2 for 14–16 days before use to allow the Caco-2 cells to differentiate. The medium in the wells was replaced by fresh medium three times a week.
Adherence assay
Overnight-grown cultures of E135, E1162 and E1162Δesp in BHI broth were diluted (1:50) and grown at 37°C to an OD660 of 0.4, while shaking. Bacteria were harvested by centrifugation (6,500 × g; 3 min) and resuspended in DMEM to a concentration of 1 × 107 CFU/ml. For each strain, 1 ml bacterial suspension was added to the wells (100 bacteria to 1 Caco-2 cell). Plates were centrifuged (175 × g; 1 min) and incubated for 1 h at 37°C in 5% CO2 to allow adherence to the Caco-2 cells. After incubation, monolayers were rinsed 3 times with DMEM and cells were lysed with 1% Triton X-100 (Merck, Darmstadt, Germany) in PBS for approximately 5 min at room temperature. Adherent bacteria were quantified by plating serial dilutions onto TSA plates and counting resultant colonies. Also the inoculum was plated to determine viable counts. The assay was performed simultaneously in 3 separate wells in duplicate and repeated on 3 different days.
Mice
Specific pathogen-free 10-week-old female C57BL/6 mice (14 mice in total) were purchased from Harlan Sprague-Dawley (Horst, The Netherlands). The animals were housed in individual cages in rooms with a controlled temperature and a 12-h light-dark cycle. They were acclimatized for 1 week prior to usage, and received standard rodent chow and water ad libitum. The Animal Care and Use Committee of the University of Amsterdam approved all experiments.
Induction of intestinal colonization
Mice were administered subcutaneous injections of ceftriaxone (Roche, Woerden, The Netherlands; 100 μl per injection, 12 mg/ml) 2 times a day, starting 2 days before inoculation of bacteria and continuing for the duration of the experiment. Two days after the initiation of the antibiotic treatment 2 × 109 CFU of E1162 or E1162Δesp in 300 μl TH broth was inoculated by orogastric inoculation using an 18-gauge stainless animal feeding tube. In addition, in one experiment mice were administered a mixture of an equal amount (1.5 × 109 CFU) of E1162 and E1162Δesp simultaneously. For all experiments, plate-grown bacteria were inoculated in TH broth and grown at 37°C to an OD620 1.0, while shaking. The inoculum was plated to determine viable counts. Mice were sacrificed after 10 days of colonization. Seven mice per group were examined.
Collection of samples
Stool samples were collected from naive mice, 2 days after antibiotic treatment and 1, 3, 6 and 10 days after bacterial inoculation. Per mice, 2 stool pellets were collected, pooled, weighed (50–129 mg), and 1 ml of sterile saline was added. After 10 days of colonization mice were anesthetized with Hypnorm® (Janssen Pharmaceutica, Beerse, Belgium; active ingredients fentanyl citrate and fluanisone) and midazolam (Roche, Meidrecht, The Netherlands), blood was drawn by cardiac puncture and transferred to heparin-gel vacutainer tubes. Mesenteric lymph nodes (MLN) were excised, weighed and collected in 4 volumes of sterile saline. Subsequently, the intestines were excised, opened and fecal contents of small bowel, cecum, and colon were weighed and 1 ml of sterile saline was added.
Determination of bacterial outgrowth
The number of E. faecium CFU was determined in stool, MLN, blood, and fecal contents of small bowel, cecum, and colon. Stool, MLN, and fecal contents were homogenized at 4°C using a tissue homogenizer (Biospec Products, Bartlesville, UK). CFU were determined from serial dilutions of the homogenates and undiluted blood. Twenty μl of each dilution and 50 μl of undiluted blood, was plated onto SB agar plates and grown at 37°C for 44 h with 5% CO2. Colonies were counted, tested by PCR to confirm species identity, and corrected for the dilution factor to calculate CFU per gram of stool/MLN/fecal contents. MLVA was performed to confirm strain identity.
PCR analysis to confirm species
Stool samples from naïve mice and from mice treated for 2 days with ceftriaxone were examined for presence of E. faecium. The lowest dilutions of stool homogenates that contained well-separated colonies were chosen and each colony of that dilution (12–24 CFU/20 μl diluted stool homogenate) was tested by PCR for presence of the housekeeping gene ddl (encoding D-alanine, D-alanine ligase) using the E. faecium specific primers ddlF (5'-GAG ACA TTG AAT ATG CCT) and ddlR (5'-AAA AAG AAA TCG CAC CG) [43]. The colonies were directly diluted in 25-μl-volumes with HotStarTaq Master Mix (QIAQEN Inc., Valencia, CA). PCR's were performed with a 9800 Fast Thermal Cycler (Applied Biosystems, Foster City, CA) and the PCR amplification conditions were as follows: initial denaturation at 95°C for 15 min, followed by 10 touchdown cycles starting at 94°C for 30 s, 60°C for 30 s, and 72°C (the time depended on the size of the PCR product) with the annealing temperature decreasing by 1°C per cycle, followed by 25 cycles with an annealing temperature of 52°C. All primers used in this study were purchased from Isogen Life Science (IJselstijn, The Netherlands).
For mono infection, colonies obtained from stool (1, 3, 6, and 10 days after bacterial inoculation), MLN, and fecal contents from small bowel, cecum, and colon were examined to confirm species identity. Colonies were randomly picked and presence of the ddl gene, in case E1162 was inoculated, or the cat gene, in case E1162Δesp was inoculated, was assessed by PCR using primer pairs ddlF – ddlR and CmF (5'-GAA TGA CTT CAA AGA GTT TTA TG) – CmR (5'-AAA GCA TTT TCA GGT ATA GGT G) [21], respectively. When both strains were inoculated simultaneously, all colonies from the lowest dilution with well-separated colonies were picked (3–28 CFU/20 μl diluted homogenate). Species identity and the number of E1162 and E1162Δesp were determined by multiplex PCR using primer pairs ddlF – ddlR and CmF – CmR. In PCR's, a colony of E1162 and E1162Δesp was used as positive control and a colony of E. faecalis V583 [44] was used as negative control.
MLVA to confirm strain identity
For both mono infection and mixed infection, colonies obtained from stool (1, 3, 6, and 10 days after bacterial inoculation), MLN, and fecal contents from small bowel, cecum, and colon were randomly picked and MLVA was performed to confirm strain identity. MLVA was performed as described previously [45].
Histological examination
Small bowel, cecum and colon tissue were fixed in 4% buffered formalin and embedded in paraffin. Four-micrometer-thick sections were stained with hematoxylin-eosin and analyzed.
Statistical analysis
Adherence data are expressed as the mean CFU per ml ± the standard deviation (SD). A two-tailed Student's t test was applied. Mouse colonization data are expressed as medians of CFU per gram of stool/fecal contents. Two group comparisons were done by Mann-Whitney U test. A p-value < 0.05 was considered statistically significant.
Abbreviations
- Esp:
-
enterococcal surface protein
- GI tract:
-
gastrointestinal tract
- Caco-2 cells:
-
human colorectal adenocarcinoma cells
- MLN:
-
mesenteric lymph nodes
- CFU:
-
colony forming units
- PCR:
-
polymerase chain reaction
- MLVA:
-
Multiple-Locus Variable-Number Tandem Repeat Analysis.
References
Murray BE: Vancomycin-resistant enterococcal infections. N Engl J Med. 2000, 342: 710-721. 10.1056/NEJM200003093421007.
Dautle MP, Ulrich RL, Hughes TA: Typing and subtyping of 83 clinical isolates purified from surgically implanted silicone feeding tubes by random amplified polymorphic DNA amplification. J Clin Microbiol. 2002, 40: 414-421. 10.1128/JCM.40.2.414-421.2002.
Edmond MB, Ober JF, Dawson JD, Weinbaum DL, Wenzel RP: Vancomycin-resistant enterococcal bacteremia: natural history and attributable mortality. Clin Infect Dis. 1996, 23: 1234-1239.
Giannitsioti E, Skiadas I, Antoniadou A, Tsiodras S, Kanavos K, Triantafyllidi H, Giamarellou H: Nosocomial vs. community-acquired infective endocarditis in Greece: changing epidemiological profile and mortality risk. Clin Microbiol Infect. 2007, 13: 763-769. 10.1111/j.1469-0691.2007.01746.x.
Leung JW, Liu YL, Desta TD, Libby ED, Inciardi JF, Lam K: In vitro evaluation of antibiotic prophylaxis in the prevention of biliary stent blockage. Gastrointest Endosc. 2000, 51: 296-303. 10.1016/S0016-5107(00)70358-0.
McDonald JR, Olaison L, Anderson DJ, Hoen B, Miro JM, Eykyn S, Abrutyn E, Fowler VG, Habib G, Selton-Suty C, Pappas PA, Cabell CH, Corey GR, Marco F, Sexton DJ: Enterococcal endocarditis: 107 cases from the international collaboration on endocarditis merged database. Am J Med. 2005, 118: 759-766. 10.1016/j.amjmed.2005.02.020.
Morrison AJ, Wenzel RP: Nosocomial urinary tract infections due to Enterococcus. Ten years' experience at a university hospital. Arch Intern Med. 1986, 146: 1549-1551. 10.1001/archinte.146.8.1549.
Mylonakis E, Calderwood SB: Infective endocarditis in adults. N Engl J Med. 2001, 345: 1318-1330. 10.1056/NEJMra010082.
Sabbuba N, Hughes G, Stickler DJ: The migration of Proteus mirabilis and other urinary tract pathogens over Foley catheters. BJU Int. 2002, 89: 55-60. 10.1046/j.1464-410X.2002.02560.x.
Svanborg C, Godaly G: Bacterial virulence in urinary tract infection. Infect Dis Clin North Am. 1997, 11: 513-529. 10.1016/S0891-5520(05)70371-8.
Tannock GW, Cook G: Enterococci as members of the intestinal microflora of humans. The enterococci: pathogenesis, molecular biology, and antibiotic resistance. Edited by: Gilmore MS, Clewell DB, Courvalin P, Dunny GM, Murray BE, Rice LBe. 2002, Washington, D.C.: American Society for Microbiology, 101-132.
Iwen PC, Kelly DM, Linder J, Hinrichs SH, Dominguez EA, Rupp ME, Patil KD: Change in prevalence and antibiotic resistance of Enterococcus species isolated from blood cultures over an 8-year period. Antimicrob Agents Chemother. 1997, 41: 494-495.
Top J, Willems RJ, Blok H, de Regt MJ, Jalink K, Troelstra A, Goorhuis B, Bonten MJ: Ecological replacement of Enterococcus faecalis by multiresistant clonal complex 17 Enterococcus faecium. Clin Microbiol Infect. 2007, 13: 316-319. 10.1111/j.1469-0691.2006.01631.x.
Treitman AN, Yarnold PR, Warren J, Noskin GA: Emerging incidence of Enterococcus faecium among hospital isolates (1993 to 2002). J Clin Microbiol. 2005, 43: 462-463. 10.1128/JCM.43.1.462-463.2005.
de Regt MJ, Wagen van der LE, Top J, Blok HE, Hopmans TE, Dekker AW, Hene RJ, Siersema PD, Willems RJ, Bonten MJ: High acquisition and environmental contamination rates of CC17 ampicillin-resistant Enterococcus faecium in a Dutch hospital. J Antimicrob Chemother. 2008, 62: 1401-1406. 10.1093/jac/dkn390.
Willems RJ, Top J, van Santen M, Robinson DA, Coque TM, Baquero F, Grundmann H, Bonten MJ: Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg Infect Dis. 2005, 11: 821-828.
Moreno F, Grota P, Crisp C, Magnon K, Melcher GP, Jorgensen JH, Patterson JE: Clinical and molecular epidemiology of vancomycin-resistant Enterococcus faecium during its emergence in a city in southern Texas. Clin Infect Dis. 1995, 21: 1234-1237.
Wells CL, Juni BA, Cameron SB, Mason KR, Dunn DL, Ferrieri P, Rhame FS: Stool carriage, clinical isolation, and mortality during an outbreak of vancomycin-resistant enterococci in hospitalized medical and/or surgical patients. Clin Infect Dis. 1995, 21: 45-50.
Leavis H, Top J, Shankar N, Borgen K, Bonten M, van Embden JD, Willems RJ: A novel putative enterococcal pathogenicity island linked to the esp virulence gene of Enterococcus faecium and associated with epidemicity. J Bacteriol. 2004, 186: 672-682. 10.1128/JB.186.3.672-682.2004.
Willems RJ, Homan W, Top J, van Santen-Verheuvel M, Tribe D, Manzioros X, Gaillard C, Vandenbroucke-Grauls CM, Mascini EM, van Kregten E, van Embden JD, Bonten MJ: Variant esp gene as a marker of a distinct genetic lineage of vancomycin-resistant Enterococcus faecium spreading in hospitals. Lancet. 2001, 357: 853-855. 10.1016/S0140-6736(00)04205-7.
Heikens E, Bonten MJ, Willems RJ: Enterococcal Surface Protein Esp is Important for Biofilm Formation of Enterococcus faecium E1162. J Bacteriol. 2007, 189: 8233-8240. 10.1128/JB.01205-07.
Van Wamel WJ, Hendrickx AP, Bonten MJ, Top J, Posthuma G, Willems RJ: Growth condition-dependent Esp expression by Enterococcus faecium affects initial adherence and biofilm formation. Infect Immun. 2007, 75: 924-931. 10.1128/IAI.00941-06.
Lund B, Edlund C: Bloodstream isolates of Enterococcus faecium enriched with the enterococcal surface protein gene, esp, show increased adhesion to eukaryotic cells. J Clin Microbiol. 2003, 41: 5183-5185. 10.1128/JCM.41.11.5183-5185.2003.
Dworniczek E, Wojciech L, Sobieszczanska B, Seniuk A: Virulence of Enterococcus isolates collected in Lower Silesia (Poland). Scand J Infect Dis. 2005, 37: 630-636. 10.1080/00365540510031421.
Shankar N, Lockatell CV, Baghdayan AS, Drachenberg C, Gilmore MS, Johnson DE: Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect Immun. 2001, 69: 4366-4372. 10.1128/IAI.69.7.4366-4372.2001.
Shankar N, Baghdayan AS, Gilmore MS: Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature. 2002, 417: 746-750. 10.1038/nature00802.
Pultz NJ, Shankar N, Baghdayan AS, Donskey CJ: Enterococcal surface protein Esp does not facilitate intestinal colonization or translocation of Enterococcus faecalis in clindamycin-treated mice. FEMS Microbiol Lett. 2005, 242: 217-219. 10.1016/j.femsle.2004.11.006.
Di Rosa R, Creti R, Venditti M, D'Amelio R, Arciola CR, Montanaro L, Baldassarri L: Relationship between biofilm formation, the enterococcal surface protein (Esp) and gelatinase in clinical isolates of Enterococcus faecalis and Enterococcus faecium. FEMS Microbiol Lett. 2006, 256: 145-150. 10.1111/j.1574-6968.2006.00112.x.
Kristich CJ, Li YH, Cvitkovitch DG, Dunny GM: Esp-independent biofilm formation by Enterococcus faecalis. J Bacteriol. 2004, 186: 154-163. 10.1128/JB.186.1.154-163.2004.
Tendolkar PM, Baghdayan AS, Gilmore MS, Shankar N: Enterococcal surface protein, Esp, enhances biofilm formation by Enterococcus faecalis. Infect Immun. 2004, 72: 6032-6039. 10.1128/IAI.72.10.6032-6039.2004.
Toledo-Arana A, Valle J, Solano C, Arrizubieta MJ, Cucarella C, Lamata M, Amorena B, Leiva J, Penades JR, Lasa I: The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl Environ Microbiol. 2001, 67: 4538-4545. 10.1128/AEM.67.10.4538-4545.2001.
Hancock LE, Perego M: The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J Bacteriol. 2004, 186: 5629-5639. 10.1128/JB.186.17.5629-5639.2004.
Hufnagel M, Koch S, Creti R, Baldassarri L, Huebner J: A putative sugar-binding transcriptional regulator in a novel gene locus in Enterococcus faecalis contributes to production of biofilm and prolonged bacteremia in mice. J Infect Dis. 2004, 189: 420-430. 10.1086/381150.
Tendolkar PM, Baghdayan AS, Shankar N: Putative surface proteins encoded within a novel transferable locus confer a high-biofilm phenotype to Enterococcus faecalis. J Bacteriol. 2006, 188: 2063-2072. 10.1128/JB.188.6.2063-2072.2006.
Kuehnert MJ, Jernigan JA, Pullen AL, Rimland D, Jarvis WR: Association between mucositis severity and vancomycin-resistant enterococcal bloodstream infection in hospitalized cancer patients. Infect Control Hosp Epidemiol. 1999, 20: 660-663. 10.1086/501561.
Matar MJ, Safdar A, Rolston KV: Relationship of colonization with vancomycin-resistant enterococci and risk of systemic infection in patients with cancer. Clin Infect Dis. 2006, 42: 1506-1507. 10.1086/503675.
Lakticova V, Hutton-Thomas R, Meyer M, Gurkan E, Rice LB: Antibiotic-induced enterococcal expansion in the mouse intestine occurs throughout the small bowel and correlates poorly with suppression of competing flora. Antimicrob Agents Chemother. 2006, 50: 3117-3123. 10.1128/AAC.00125-06.
Pultz NJ, Stiefel U, Subramanyan S, Helfand MS, Donskey CJ: Mechanisms by which anaerobic microbiota inhibit the establishment in mice of intestinal colonization by vancomycin-resistant Enterococcus. J Infect Dis. 2005, 191: 949-956. 10.1086/428090.
Pultz NJ, Vesterlund S, Ouwehand AC, Donskey CJ: Adhesion of vancomycin-resistant Enterococcus to human intestinal mucus. Curr Microbiol. 2006, 52: 221-224. 10.1007/s00284-005-0244-2.
Leavis HL, Willems RJ, Van Wamel WJ, Schuren FH, Caspers MP, Bonten MJ: Insertion sequence-driven diversification creates a globally dispersed emerging multiresistant subspecies of Enterococcus faecium. PLoS Pathog. 2007, 3: e7-10.1371/journal.ppat.0030007.
Hendrickx AP, Van Wamel WJ, Posthuma G, Bonten MJ, Willems RJ: Five genes encoding surface-exposed LPXTG proteins are enriched in hospital-adapted Enterococcus faecium clonal complex 17 isolates. J Bacteriol. 2007, 189: 8321-8332. 10.1128/JB.00664-07.
Heikens E, van Schaik W, Leavis HL, Bonten MJ, Willems RJ: Identification of a novel genomic island specific to hospital-acquired clonal complex 17 Enterococcus faecium isolates. Appl Environ Microbiol. 2008, 74: 7094-7097. 10.1128/AEM.01378-08.
Ozawa Y, Courvalin P, Gaiimand M: Identification of enterococci at the species level by sequencing of the genes for D-alanine:D-alanine ligases. Syst Appl Microbiol. 2000, 23: 230-237.
Sahm DF, Kissinger J, Gilmore MS, Murray PR, Mulder R, Solliday J, Clarke B: In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother. 1989, 33: 1588-1591.
Top J, Schouls LM, Bonten MJ, Willems RJ: Multiple-locus variable-number tandem repeat analysis, a novel typing scheme to study the genetic relatedness and epidemiology of Enterococcus faecium isolates. J Clin Microbiol. 2004, 42: 4503-4511. 10.1128/JCM.42.10.4503-4511.2004.
Acknowledgements
This work was supported by the European Union Sixth Framework Programme "Approaches to Control multi-resistant Enterococci (ACE): Studies on molecular ecology, horizontal gene transfer, fitness and prevention" under contract LSHE-CT-2007-037410 and ZonMW "Vaccine-development to combat the emergence of vancomycin-resistant Enterococcus faecium" project number 0.6100.0008. The authors thank J. Daalhuisen and M. ten Brink for their expert technical assistance and E. Duizer for helpful comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
EH and ML carried out the design of the study, performed the mice and cell adherence experiments, and drafted the manuscript. LMW participated in the cell adherence experiments and helped to draft the manuscript. MvLA participated in the PCR analysis to confirm species. MJMB participated in the design of the study and helped to draft the manuscript. TvdP and RJLW participated in the design and coordination of the study, and helped to draft the manuscript. All authors read and approved the final manuscript.
Esther Heikens, Masja Leendertse contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Heikens, E., Leendertse, M., Wijnands, L.M. et al. Enterococcal surface protein Esp is not essential for cell adhesion and intestinal colonization of Enterococcus faeciumin mice. BMC Microbiol 9, 19 (2009). https://doi.org/10.1186/1471-2180-9-19
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2180-9-19