Abstract
Background
Paracoccidioides brasiliensis is a dimorphic fungus that causes the most prevalent systemic mycosis in Latin America. The response to heat shock is involved in pathogenesis, as this pathogen switches from mycelium to yeast forms in a temperature dependent fashion that is essential to establish infection. HSP90 is a molecular chaperone that helps in the folding and stabilization of selected polypeptides. HSP90 family members have been shown to present important roles in fungi, especially in the pathogenic species, as an immunodominant antigen and also as a potential antifungal therapeutic target.
Results
In this work, we decided to further study the Pbhsp90 gene, its expression and role in cell viability because it plays important roles in fungal physiology and pathogenesis. Thus, we have sequenced a Pbhsp90 cDNA and shown that this gene is present on the genome as a single copy. We have also confirmed its preferential expression in the yeast phase and its overexpression during dimorphic transition and oxidative stress. Treatment of the yeast with the specific HSP90 inhibitors geldanamycin and radicicol inhibited growth at 2 and 10 μM, respectively.
Conclusion
The data confirm that the Pbhsp90 gene encodes a morphologically regulated and stress-responsive protein whose function is essential to cell viability of this pathogen. This work also enforces the potential of HSP90 as a target for antifungal therapies, since the use of HSP90 inhibitors is lethal to the P. brasiliensis yeast cells in a dose-responsive manner.
Similar content being viewed by others
Background
Most cells react to temperature elevations in a stereotypical manner termed heat shock response, which is conserved from bacteria to mammals [1]. Upon heating, the cell almost completely represses transcription and translation except for a set of so-called heat shock proteins (HSPs), thus initiating its adaptation to the new environment. The induction of molecular chaperones is also seen after non-thermal stresses such as those caused by oxidative stress, low pH and treatment with cytotoxic drugs.
HSP90 is a molecular chaperone with classical in vitro activity of protein folding. However, unlike other molecular chaperones, in vivo HSP90 is not necessary for de novo protein synthesis; it assists only a small set of proteins, which are usually dependent on ATP-dependent HSP90 binding to perform their functions correctly [2]. HSP90 client proteins include several tyrosine and serine/threonine kinases, steroid receptors and transcription factors [3]. The HSP90 protein consists of a highly conserved N-terminal nucleotide-binding domain, a flexible charged linker and a C-terminal domain that contains a -MEEVD conserved motif responsible for binding to tetratricopeptide-repeat (TPR) proteins [4].
HSP90 family members have been shown to play important roles in fungi. In the model yeast Saccharomyces cerevisiae, inhibition of HSP90 by an anti-HSP90 ribozyme promotes cell lysis, indicating some potential as an antifungal therapeutic target [5]. The Candida albicans HSP90 has been shown to be an immunodominant antigen, both in a mouse model of infection and in human patients [6]. Based on this finding, a novel therapeutic strategy has been devised using a human recombinant antibody to HSP90, which shows intrinsic antifungal activity and synergy with amphotericin B both in vitro and in vivo. This antibody is in clinical trials with encouraging results against systemic candidiasis [7]. HSP90 immunogenicity has also been applied to a phage-displayed vaccine tested in mice, which acquired resistance to systemic C. albicans infection [8].
HSP90 proteins are targeted by several different pharmacologic agents; two of them, radicicol and geldanamycin, inhibit the protein's ATPase activity with great specificity and potency. Derivatives of these drugs and several novel HSP90 inhibitors are now in clinical trials for cancer chemotherapy [9]. However, cancer chemotherapy is not the only use for these drugs, as they have become useful in studies of HSP90 function. Inactivation of the Leishmania donovani HSP90 with geldanamycin or radicicol mimics the transmission from the vector to the mammalian host, inducing the differentiation from the insect promastigote stage towards the pathogenic mammalian amastigote stage [10]. HSP90 also influences stage differentiation in Toxoplasma gondii, which led to its proposition as a potential drug target [11].
Paracoccidioides brasiliensis, a dimorphic fungus, can be found either as a filamentous soil saprobe or as yeast cells infecting mammalian hosts. Both forms can be cultivated in vitro in different incubating temperatures, around 22°C for mycelium and 36°C for yeast. Mycelium cells can be transformed into yeast by raising the incubation temperature to 36°C; and then reverted to mycelium by lowering the temperature [12]. The fungus undergoes a similar morphological switch in vivo: the infection starts by inhalation of conidia or mycelium fragments from the environment, which then transform into yeast in the host's lungs. Pathogenicity has been intimately associated with this process, as strains unable to differentiate into the yeast form are avirulent [13]. Adaptation to different temperatures thus seems to be paramount to both fungal physiology and pathogenicity, a fact that has long elicited interest on heat shock response in such pathogens. Several P. brasiliensis molecular chaperones have been studied, such as MDJ1 [14], HSP60 [15], HSP70 genes [16–19] and HSP100 genes [20, 21]. A boost in the knowledge of these genes came from high throughput transcriptome sequencing and expression analyses [22–24], which revealed cDNA sequences and some expression data from 48 molecular chaperones [25].
This work describes the complete cDNA characterization of the HSP90 gene from P. brasiliensis (Pbhsp90) and its expression profile, including differential expression in yeast phase, induction during mycelium to yeast transition and oxidative stress. Moreover, analysis of its role on cell viability was investigated with pharmacological inhibitors.
Results and Discussion
The Pbhsp90 cDNA was first isolated during the P. brasiliensis transcriptome sequencing [22, 24]. In order to confirm the hsp90 transcript sequence, we chose a single clone which clustered in the Pbhsp90 transcriptome contig and fully sequenced it. The sequence shows a single open reading frame (ORF) encoding a predicted polypeptide with 706 amino acid residues that aligned with several known HSP90 proteins from organisms as diverse as fungi, plants and mammals. A search for conserved domains using NCBI rpsblast identified the Bergerat-fold N-terminal ATPase domain [26] that is typical of all HSP90 proteins. Nine out of ten residues involved in ATP/ADP and geldanamycin binding at the N-terminal domain (Leu-34, Asn-37, Lys-44, Asp-79, Gly-83, Met-84, Asn-92, Phe-124 and Thr-171) were identical in P. brasiliensis and S. cerevisiae, the only difference being a change from a lysine to an arginine at position 98 [27]. The predicted PbHSP90 protein also presents the C-terminal conserved -MEEVD motif. A total of 400 out of the 706 amino acid residues are conserved across Homo sapiens, S. cerevisiae and P. brasiliensis (figure 1).
ClustalW alignment of PbHSP90 and other HSP90 family proteins. The Pbhsp90 ORF was translated and aligned with Saccharomyces cerevisiae (Hsp82p, accession number [GenBank: NP_015084.1]) and Homo sapiens (HSP90β, accession number [GenBank: AAQ63401.1]) homologues using ClustalW. Shaded amino acid residues are involved in the binding of either ATP/ADP or geldanamycin in S. cerevisiae [27]. The N-terminal MEEVD motif (bold) is the binding site for the TPR domain of co-chaperones. A total of 400 out of the 706 amino acid residues are conserved across the three species.
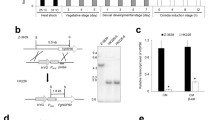
All prokaryotes and some eukaryotes are known to have a single cytoplasmic copy of the hsp90 gene, but some fungi present two copies [28, 29]. In the later organisms, the two genes encode proteins which are structurally very similar and functionally identical. The only significant difference between the two genes is their transcriptional pattern; one is constitutively expressed and mildly induced during stress, while the other is markedly stress-induced [28]. The expression of an HSP90 gene from an organism that harbours a single copy, e.g. C. albicans, is somewhat like a combination of both patterns, with constitutive expression and marked stress induction of the same gene [29]. The next step in characterizing the Pbhsp90 gene was to evaluate the number of copies present in the genome. Southern blotting analysis using the Pbhsp90 cDNA as a probe has shown only one copy on the genome (Figure 2). In addition, searching the recently released genome sequence drafts from isolates Pb01, Pb18 and Pb03 [30] with the Pbhsp90 cDNA reveals a single copy of the gene. The translated sequence in these genomes contains three conserved introns and the predicted proteins are 98–99% identical to the one we obtained in this work.
Southern blot analysis of gene copy number. Samples of 15 μg total DNA were digested with combinations of the restriction endonucleases ApaI (A), NdeI (N) and SacII (S), separated by agarose gel electrophoresis, blotted onto charged nylon membrane and hybridized with radioactively labelled PbHSP90 cDNA. Bars show molecular weight marker.
The Pbhsp90 gene expression was then assessed by northern blotting under several different experimental conditions including non-stressed mycelium and yeast cultures, cells undergoing the first 24 hours of the mycelium-to-yeast transition, and oxidative-stressed yeast cells (Figure 3). As previously shown by electronic subtraction, cDNA microarray and real-time RT-PCR [22–24], expression of the Pbhsp90 gene can be observed in both morphological phases, but it is around 4.5 times stronger in the yeast phase (Figure 3 – panel A). These results are in accordance to what has been observed in another dimorphic fungus, Penicillium marneffei, in which the HSP90 protein is 5.29 times more expressed in the yeast phase [31]. The mycelium mRNA is slightly shorter than the yeast one, probably due to differences in the 3'- and/or 5'-untranslated regions (UTRs) as ESTs from mycelium and yeast formed a single cluster which corresponds to only one transcript; this has been described before for other fungal genes [32]. Pbhsp90 expression during the mycelium-to-yeast transition (Figure 3 – panel B) is already increased 30 minutes after the temperature shift, reaches its peak – about 25-fold relative induction – at one hour and slowly decreases during the following hours, reflecting a morphological transition thermo-dependent response [33], which is also observed by EST analysis [34]. A very similar pattern of induction has been shown for the chaperone-encoding genes hsp60 [15], hsp70 [16, 18] and hsp104 [23].
Pbhsp90 expression profiles in mycelium and yeast cells, during differentiation and in the presence of oxidative agents. Total RNA (15 μg) extracted from P. brasiliensis was separated by denaturing agarose gel electrophoresis, blotted onto charged nylon membranes and probed with radioactively labelled Pbhsp90 cDNA. Panel A – mycelium (M) and yeast (Y) cells. Panel B – Mycelium cells grown at 22°C and after incubation at 36°C for up to 24 h. The membrane shown on panel B was washed to higher stringency to resolve the strong hybridization signals better. Panel C – Yeast cells incubated with 15 mM hydrogen peroxide or 1 mM menadione, a superoxide-generating reagent. In each panel, the top image represents the specific hybridization signal and the ethidium bromide stained 16S rRNA band in the bottom image.
HSP90 is known to be essential for the function of several signal transduction kinases in organisms as diverse as fruit flies [35], baker's yeast [36] and mammalians [37], including members of the MAPK and PKA families involved in fungal morphogenesis [38]. These proteins, as most in the cell after a temperature elevation of more than 10°C, are prone to denature under circumstances when signalling must occur to coordinate the transition. The very large increase on the Pbhsp90 mRNA levels may thus be seen not only as result of heat shock, but also as adaptation in the transition.
Another set of northern blots examined Pbhsp90 gene expression under some of the conditions previously shown to induce other fungal homologues. Hydrogen peroxide treatment promoted only a slight increase in Pbhsp90 expression, of 2.45-fold after 30 minutes (Figure 3, panel C). The H2O2 concentration used (15 mM) is far below the maximum tolerated by P. brasiliensis, which is close to 75 mM [39], but is three times higher than the LD90 for S. cerevisiae cells at 30 minutes [40]. Differently, two other chaperones have been shown to be induced by 5 mM H2O2 in P. brasiliensis [14]. In contrast to the poor induction provoked by peroxide on Pbhsp90, treatment with superoxide-generating menadione at 1 mM (Figure 3 – panel C) strongly induced Pbhsp90 expression in yeast cells. H2O2 and superoxide are among the major reactive oxygen species (ROS) used by mammalian macrophages in the defence against fungal infection [41]. C. albicans cells lacking Cu/Zn superoxide dismutase (SOD), a superoxide detoxification enzyme, showed increased susceptibility to macrophage attack and had attenuated virulence in mice [42]. In a review of the transcriptome sequencing project, Campos et al. [43] have found three catalases and three SODs in P. brasiliensis -isolate Pb01. The fact that superoxide radicals strongly induced Pbhsp90 correlates nicely with recent data from our group showing that SOD specific activity in the P. brasiliensis yeast cell is as strong as in non-pathogenic S. cerevisiae (Dantas et al., personal communication), while specific catalase activity is over 2000 times higher [39]. Therefore, it seems that the H2O2 detoxification by P. brasiliensis is so efficient that little cellular damage requiring HSP90 occurs.
It is not possible to study Pbhsp90 function by classical genetic approaches because knockouts of members of this family are usually not viable [28]. Furthermore, genetic manipulation is still not optimized on the multinucleated and multi-budding P. brasiliensis making RNA interference unfeasible as well. Those challenges have been circumvented by means of a "pharmacological knockout" with the potent and specific HSP90 inhibitors radicicol and geldanamycin. A broth microdilution test shows that both radicicol and geldanamycin inhibit the growth of P. brasiliensis – Pb01 yeast cells in a manner similar to amphotericin B (Figure 4), meaning that both drugs are able to penetrate P. brasiliensis cells and bind their targets. Moreover, it confirms that HSP90 inhibition is lethal to the cell in a dose-responsive manner and consequently that its function is essential to fungal physiology. Results similar to these have been found in other microorganisms such as S. cerevisiae and Schizosaccharomyces pombe [5, 44]. The use of sub-lethal concentrations of these drugs can be foreseen as an excellent form of further investigating the broad heat shock response in P. brasiliensis and in special Pbhsp90.
Susceptibility of P. brasiliensis yeast cells to HSP90 inhibitors geldanamycin and radicicol. Inhibition of cell growth by HSP90 targeting drugs or the control amphotericin B was tested by a broth microdilution test adapted from the international standard M27-A2. Curves show mean plus standard error of the absorbance at OD595 nm measured from quadruplicate experiments. *statistically significant when compared to control (p < 0.05).
Conclusion
The data show that Pbhsp90 is a highly expressed gene, under complex regulation by morphological and oxidative stress signals. This picture resembles previous observations of other molecular chaperones, stressing even more the importance of heat adaptation in P. brasiliensis dimorphism and survival in the harsh environment inside the mammalian host. This work also enforces the potential of HSP90 as a target for novel antifungal therapies, since the use of HSP90 inhibitors is lethal to the P. brasiliensis yeast cells in a dose-responsive manner. Geldanamycin, radicicol and the other members of the ever-growing list of HSP90 inhibitors could be engineered to achieve higher selectiveness for the fungal protein. Taken together, these results confirm that the Pbhsp90 gene encodes a morphologically and stress-responsive protein whose function is essential to cell viability of this pathogen.
Methods
Strains and cultures
P. brasiliensis strain Pb01 (ATCC-MYA-826) was used throughout this study. Cells were maintained on semi-solid Fava-Netto medium, incubated at 37°C for yeast cells and 22°C for mycelia.
cDNA sequencing and assembly
A single cDNA cloned in a Lambda ZAP phage vector (Stratagene) was excised in vivo according to the supplier's manual. The resulting plasmid was sequenced with vector-encoded T3 and T7 and internal primers in a MegaBACE® (GE Healthcare) automatic sequencer. Base-calling, filtering and assembly were done as described before [24]. The sequence was deposited in GenBank (accession number [GenBank: AY928608]).
Southern blot
Total DNA was obtained by phenol:chloroform extraction of mechanically disrupted frozen yeast cells. Samples of 15 μg of DNA were digested, with combinations of three restriction endonucleases (ApaI, NdeI and SacII) known not to have internal sites in the Pbhsp90 cDNA, and separated by agarose gel electrophoresis. The gel was blotted onto a charged nylon membrane by upward capillary transfer. The cDNA probe was amplified by PCR using vector primers. A total of 25 ng of the purified product was radioactively labelled with α-P32-dATP (GE Healthcare) using the MegaPrime labelling kit (GE Healthcare). The hybridized membrane was then washed as described for the northern blots below sequentially until low background was detected and then exposed to a phosphorimager, which was scanned on a Typhoon® 9210 scanner (GE Healthcare).
Northern blot
RNAs from mycelium-to-yeast transition and of isolated mycelium and yeast cells were obtained as previously described [45, 46]. For the oxidative stress experiments, yeast cells were washed and ressuspended in medium containing either 15 mM H2O2 or 1 mM menadione. Samples were then collected by centrifugation, frozen with liquid nitrogen and disrupted. Total RNA was then obtained by extraction with Trizol® reagent (Invitrogen). Fifteen microgram samples of total RNA were separated by electrophoresis on formaldehyde-containing 1% agarose gels; blotted onto charged nylon Hybond® N+ membranes (GE Healthcare) and hybridized as described for the Southern-blot. Hybridized membranes were then washed sequentially with solutions containing SDS 0.1% and different concentrations of SSPE in 20 minute rounds: 2× SSPE 50°C, 1× SSPE 50°C, 1× SSPE 65°C and 0,1× SSPE 65°C. Each membrane was monitored with a Geiger counter after each wash and exposed in a phosphorimager when the background radioactivity was low enough. Resulting bands were quantified using ImageQuant® software and normalized by dividing the hybridization signal intensity by the ethidium bromide stained 18S ribosomal RNA band intensity.
Drug susceptibility testing
Drug susceptibility tests were adapted from the international standard M27-A2 [47]. Yeast cells were grown on RPMI defined medium and diluted to a 2× suspension containing 105 cells/mL. Amphotericin B and geldanamycin were diluted in dimethyl-sulfoxide (DMSO) to 100× stock solutions; radicicol was diluted in ethanol. Working solutions (2×) were then made with RPMI medium. A 100 μL aliquot of the 2× cell suspension was added to 100 μL of the 2× drug solutions in U-shaped 96-well plates, so that the final suspension reached a cell density of 105 cells/mL and 1% DMSO or ethanol. The plate was incubated at 37°C for 7–10 days, when the minimal inhibitory concentration (MIC) was directly observed. In order to convey more precisely the observed difference in growth, OD595 nm was measured with a GeneQuant® spectrophotometer. Each concentration was tested in quadruplicate experiments. Statistical analyses were done with one-way analysis of variance (ANOVA) and the Tamhane T2 multiple comparison test from SPSS 11.0 software.
References
Walter S, Buchner J: Molecular chaperones – cellular machines for protein folding. Angew Chem Int Ed Engl. 2002, 41 (7): 1098-1113. 10.1002/1521-3773(20020402)41:7<1098::AID-ANIE1098>3.0.CO;2-9.
Nathan DF, Vos MH, Lindquist S: In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc Natl Acad Sci USA. 1997, 94 (24): 12949-12956. 10.1073/pnas.94.24.12949.
Buchner J: Hsp90 & Co. – a holding for folding. Trends Biochem Sci. 1999, 24 (4): 136-141. 10.1016/S0968-0004(99)01373-0.
Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I: Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000, 101 (2): 199-210. 10.1016/S0092-8674(00)80830-2.
Sreedhar AS, Mihaly K, Pato B, Schnaider T, Stetak A, Kis-Petik K, Fidy J, Simonics T, Maraz A, Csermely P: Hsp90 inhibition accelerates cell lysis. Anti-Hsp90 ribozyme reveals a complex mechanism of Hsp90 inhibitors involving both superoxide- and Hsp90-dependent events. J Biol Chem. 2003, 278 (37): 35231-35240. 10.1074/jbc.M301371200.
Matthews R, Burnie J: Cloning of a DNA sequence encoding a major fragment of the 47 kilodalton stress protein homologue of Candida albicans. FEMS Microbiol Lett. 1989, 51 (1): 25-30. 10.1016/0378-1097(89)90071-2.
Pachl J, Svoboda P, Jacobs F, Vandewoude K, Hoven van der B, Spronk P, Masterson G, Malbrain M, Aoun M, Garbino J, Takala J, Drgona L, Burnie J, Matthews R, Mycograb Invasive Candidiasis Study Group: A randomized, blinded, multicenter trial of lipid-associated amphotericin B alone versus in combination with an antibody-based inhibitor of heat shock protein 90 in patients with invasive candidiasis. Clin Infect Dis. 2006, 42 (10): 1404-1413. 10.1086/503428.
Yang Q, Wang L, Lu DN, Gao RJ, Song JN, Hua PY, Yuan DW: Prophylactic vaccination with phage-displayed epitope of C. albicans elicits protective immune responses against systemic candidiasis in C57BL/6 mice. Vaccine. 2005, 23 (31): 4088-4096. 10.1016/j.vaccine.2004.07.005.
Miyata Y: Hsp90 inhibitor geldanamycin and its derivatives as novel cancer chemotherapeutic agents. Curr Pharm Des. 2005, 11 (9): 1131-1138. 10.2174/1381612053507585.
Wiesgigl M, Clos J: Heat shock protein 90 homeostasis controls stage differentiation in Leishmania donovani. Mol Biol Cell. 2001, 12 (11): 3307-3316.
Echeverria PC, Matrajt M, Harb OS, Zappia MP, Costas MA, Roos DS, Dubremetz JF, Angel SO: Toxoplasma gondii Hsp90 is a potential drug target whose expression and subcellular localization are developmentally regulated. J Mol Biol. 2005, 350 (4): 723-734. 10.1016/j.jmb.2005.05.031.
San-Blas G, Nino-Vega G, Iturriaga T: Paracoccidioides brasiliensis and paracoccidioidomycosis: molecular approaches to morphogenesis, diagnosis, epidemiology, taxonomy and genetics. Med Mycol. 2002, 40 (3): 225-242. 10.1080/714031110.
Hallak J, San-Blas F, San-Blas G: Isolation and wall analysis of dimorphic mutants of Paracoccidioides brasiliensis. Sabouraudia. 1982, 20 (1): 51-62.
Batista WL, Barros TF, Goldman GH, Morais FV, Puccia R: Identification of transcription elements in the 5' intergenic region shared by LON and MDJ1 heat shock genes from the human pathogen Paracoccidioides brasiliensis. Evaluation of gene expression. Fungal Genet Biol. 2007, 44 (5): 347-356. 10.1016/j.fgb.2006.11.002.
Izacc SM, Gomez FJ, Jesuino RS, Fonseca CA, Felipe MS, Deepe GS, Soares CM: Molecular cloning, characterization and expression of the heat shock protein 60 gene from the human pathogenic fungus Paracoccidioides brasiliensis. Med Mycol. 2001, 39 (5): 445-455. 10.1080/714031053.
da Silva SP, Borges-Walmsley MI, Pereira IS, Soares CM, Walmsley AR, Felipe MS: Differential expression of an hsp70 gene during transition from the mycelial to the infective yeast form of the human pathogenic fungus Paracoccidioides brasiliensis. Mol Microbiol. 1999, 31 (4): 1039-1050. 10.1046/j.1365-2958.1999.01236.x.
Diez S, Gomez BL, Restrepo A, Hay RJ, Hamilton AJ: Paracoccidioides brasiliensis 87-kilodalton antigen, a heat shock protein useful in diagnosis: characterization, purification, and detection in biopsy material via immunohistochemistry. J Clin Microbiol. 2002, 40 (2): 359-365. 10.1128/JCM.40.2.359-365.2002.
Florez AM, Oviedo A, Cardona A, Herrera M, Garcia E, Restrepo A, McEwen JG: Molecular cloning and characterization of two hsp 70 homologous genes from the dimorphic fungus Paracoccidioides brasiliensis. Biomedica. 2003, 23 (4): 424-436.
Bisio LC, Silva SP, Pereira IS, Xavier MA, Venancio EJ, Puccia R, Soares CM, Felipe MS: A new Paracoccidioides brasiliensis 70-kDa heat shock protein reacts with sera from paracoccidioidomycosis patients. Med Mycol. 2005, 43 (6): 495-503. 10.1080/13693780400029478.
Jesuino RS, Azevedo MO, Felipe MS, Pereira M, De Almeida Soares CM: Characterization of a chaperone ClpB homologue of Paracoccidioides brasiliensis. Yeast. 2002, 19 (11): 963-972. 10.1002/yea.888.
Camargos Oliveira J, Da Silva Castro N, Soares Felipe MS, Pereira M, De Almeida Soares CM: Comparative analysis of the cDNA encoding a ClpA homologue of Paracoccidioides brasiliensis. Mycol Res. 2005, 109 (Pt 6): 707-716. 10.1017/S0953756205002789.
Felipe MS, Andrade RV, Petrofeza SS, Maranhao AQ, Torres FA, Albuquerque P, Arraes FB, Arruda M, Azevedo MO, Baptista AJ, Bataus LA, Borges CL, Campos EG, Cruz MR, Daher BS, Dantas A, Ferreira MA, Ghil GV, Jesuino RS, Kyaw CM, Leitão L, Martins CR, Moraes LM, Neves EO, Nicola AM, Alves ES, Parente JA, Pereira M, Poças-Fonseca MJ, Resende R, Ribeiro BM, Saldanha RR, Santos SC, Silva-Pereira I, Silva MA, Silveira E, Simões IC, Soares RB, Souza DP, De-Souza MT, Andrade EV, Xavier MA, Veiga HP, Venancio EJ, Carvalho MJ, Oliveira AG, Inoue MK, Almeida NF, Walter ME, Soares CM, Brígido MM: Transcriptome characterization of the dimorphic and pathogenic fungus Paracoccidioides brasiliensis by EST analysis. Yeast. 2003, 20 (3): 263-271. 10.1002/yea.964.
Goldman GH, dos Reis Marques E, Duarte Ribeiro DC, de Souza Bernardes LA, Quiapin AC, Vitorelli PM, Savoldi M, Semighini CP, de Oliveira RC, Nunes LR, Travassos LR, Puccia R, Batista WL, Ferreira LE, Moreira JC, Bogossian AP, Tekaia F, Nobrega MP, Nobrega FG, Goldman MH: Expressed sequence tag analysis of the human pathogen Paracoccidioides brasiliensis yeast phase: identification of putative homologues of Candida albicans virulence and pathogenicity genes. Eukaryot Cell. 2003, 2 (1): 34-48. 10.1128/EC.2.1.34-48.2003.
Felipe MS, Andrade RV, Arraes FB, Nicola AM, Maranhao AQ, Torres FA, Silva-Pereira I, Pocas-Fonseca MJ, Campos EG, Moraes LM, Andrade PA, Tavares AH, Silva SS, Kyaw CM, Souza DP, Pereira M, Jesuíno RS, Andrade EV, Parente JA, Oliveira GS, Barbosa MS, Martins NF, Fachin AL, Cardoso RS, Passos GA, Almeida NF, Walter ME, Soares CM, Carvalho MJ, Brígido MM, PbGenome Network: Transcriptional profiles of the human pathogenic fungus Paracoccidioides brasiliensis in mycelium and yeast cells. J Biol Chem. 2005, 280 (26): 24706-24714. 10.1074/jbc.M500625200.
Nicola AM, Andrade RV, Silva-Pereira I: Molecular chaperones in the Paracoccidioides brasiliensis transcriptome. Genet Mol Res. 2005, 4 (2): 346-357.
Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P: An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997, 386 (6623): 414-417. 10.1038/386414a0.
Prodromou C, Roe SM, O'Brien R, Ladbury JE, Piper PW, Pearl LH: Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell. 1997, 90 (1): 65-75. 10.1016/S0092-8674(00)80314-1.
Borkovich KA, Farrelly FW, Finkelstein DB, Taulien J, Lindquist S: hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol Cell Biol. 1989, 9 (9): 3919-3930.
Swoboda RK, Bertram G, Budge S, Gooday GW, Gow NA, Brown AJ: Structure and regulation of the HSP90 gene from the pathogenic fungus Candida albicans. Infect Immun. 1995, 63 (11): 4506-4514.
The Paracoccidioides brasiliensis genome Database. [http://www.niaid.nih.gov/dmid/genomes/mscs/default.htm]
Xi L, Xu X, Liu W, Li X, Liu Y, Li M, Zhang J, Li M: Differentially expressed proteins of pathogenic Penicillium marneffei in yeast and mycelial phases. J Med Microbiol. 2007, 56 (Pt 3): 298-304. 10.1099/jmm.0.46808-0.
Hwang L, Hocking-Murray D, Bahrami AK, Andersson M, Rine J, Sil A: Identifying phase-specific genes in the fungal pathogen Histoplasma capsulatum using a genomic shotgun microarray. Mol Biol Cell. 2003, 14 (6): 2314-2326. 10.1091/mbc.E03-01-0027.
Morimoto RI, Sarge KD, Abravaya K: Transcriptional regulation of heat shock genes. A paradigm for inducible genomic responses. J Biol Chem. 1992, 267 (31): 21987-21990.
Bastos KP, Bailao AM, Borges CL, Faria FP, Felipe MS, Silva MG, Martins WS, Fiuza RB, Pereira M, Soares CM: The transcriptome analysis of early morphogenesis in Paracoccidioides brasiliensis mycelium reveals novel and induced genes potentially associated to the dimorphic process. BMC Microbiol. 2007, 7: 29-10.1186/1471-2180-7-29.
Cutforth T, Rubin GM: Mutations in Hsp83 and cdc37 impair signaling by the sevenless receptor tyrosine kinase in Drosophila. Cell. 1994, 77 (7): 1027-1036. 10.1016/0092-8674(94)90442-1.
Millson SH, Truman AW, King V, Prodromou C, Pearl LH, Piper PW: A two-hybrid screen of the yeast proteome for Hsp90 interactors uncovers a novel Hsp90 chaperone requirement in the activity of a stress-activated mitogen-activated protein kinase, Slt2p (Mpk1p). Eukaryot Cell. 2005, 4 (5): 849-860. 10.1128/EC.4.5.849-860.2005.
Perdew GH, Wiegand H, Heuvel Vanden JP, Mitchell C, Singh SS: A 50 kilodalton protein associated with raf and pp60(v-src) protein kinases is a mammalian homolog of the cell cycle control protein cdc37. Biochemistry. 1997, 36 (12): 3600-3607. 10.1021/bi9612529.
Dhillon NK, Sharma S, Khuller GK: Signaling through protein kinases and transcriptional regulators in Candida albicans. Crit Rev Microbiol. 2003, 29 (3): 259-275. 10.1080/713610451.
Dantas AS, Andrade RV, de Carvalho MJ, Felipe MS, Campos EG: Oxidative stress response in Paracoccidioides brasiliensis: assessing catalase and cytochrome c peroxidase. Mycol Res. 2008, 112 (Pt 6): 747-756. 10.1016/j.mycres.2007.11.018.
Izawa S, Inoue Y, Kimura A: Importance of catalase in the adaptive response to hydrogen peroxide: analysis of acatalasaemic Saccharomyces cerevisiae. Biochem J. 1996, 320 (Pt 1): 61-67.
Vazquez-Torres A, Balish E: Macrophages in resistance to candidiasis. Microbiol Mol Biol Rev. 1997, 61 (2): 170-192.
Hwang CS, Rhie GE, Oh JH, Huh WK, Yim HS, Kang SO: Copper- and zinc-containing superoxide dismutase (Cu/ZnSOD) is required for the protection of Candida albicans against oxidative stresses and the expression of its full virulence. Microbiology. 2002, 148 (Pt 11): 3705-3713.
Campos EG, Jesuino RS, Dantas Ada S, Brigido Mde M, Felipe MS: Oxidative stress response in Paracoccidioides brasiliensis. Genet Mol Res. 2005, 4 (2): 409-429.
Ki SW, Kasahara K, Kwon HJ, Ishigami K, Kitahara T, Beppu T, Yoshida M, Horinouchi S: Radicicol binding to Swo1/Hsp90 and inhibition of growth of specific temperature-sensitive cell cycle mutants of fission yeast. Biosci Biotechnol Biochem. 2001, 65 (11): 2528-2534. 10.1271/bbb.65.2528.
Venancio EJ, Kyaw CM, Mello CV, Silva SP, Soares CM, Felipe MS, Silva-Pereira I: Identification of differentially expressed transcripts in the human pathogenic fungus Paracoccidioides brasiliensis by differential display. Med Mycol. 2002, 40 (1): 45-51. 10.1080/714031083.
Andrade RV, Paes HC, Nicola AM, de Carvalho MJ, Fachin AL, Cardoso RS, Silva SS, Fernandes L, Silva SP, Donadi EA, Sakamoto-Hojo ET, Passos GA, Soares CM, Brígido MM, Felipe MS: Cell organisation, sulphur metabolism and ion transport-related genes are differentially expressed in Paracoccidioides brasiliensis mycelium and yeast cells. BMC Genomics. 2006, 7: 208-10.1186/1471-2164-7-208.
National Committee for Clinical Laboratory Standards: Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standard – Second Edition. 2002, Wayne, PA: National Committee for Clinical Laboratory Standards
Acknowledgements
The authors wish to thank Izabel da Silva for the statistical analyses and Hugo Costa Paes and Erin McClelland for text draft revision.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
AMN planned and designed the study, performed the experiments and analyzed their results and drafted the manuscript. RVA, ASD and PAA prepared samples for and executed Northern blots. ASD and FBMA participated in cDNA cloning and sequencing. LF executed drug susceptibility assays and reviewed the manuscript. ISP supervised cDNA cloning, Southern and northern blots and participated on data analysis. MSSF conceived and coordinated the study, data analysis and manuscript preparation. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Nicola, A.M., Andrade, R.V., Dantas, A.S. et al. The stress responsive and morphologically regulated hsp90 gene from Paracoccidioides brasiliensis is essential to cell viability. BMC Microbiol 8, 158 (2008). https://doi.org/10.1186/1471-2180-8-158
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2180-8-158