Abstract
Background
Human myelogenous leukemia characterized by either the non random t(8; 21)(q22; q22) or t(16; 21)(q24; q22) chromosome translocations differ for both their biological and clinical features. Some of these features could be consequent to differential epigenetic transcriptional deregulation at AML1 targets imposed by AML1-MTG8 and AML1-MTG16, the fusion proteins deriving from the two translocations. Preliminary findings showing that these fusion proteins lead to transcriptional downregulation of AML1 targets, marked by repressive chromatin changes, would support this hypothesis. Here we show that combining conventional global gene expression arrays with the power of bioinformatic genomic survey of AML1-consensus sequences is an effective strategy to identify AML1 targets whose transcription is epigenetically downregulated by the leukemia-associated AML1-MTG16 protein.
Results
We interrogated mouse gene expression microarrays with probes generated either from 32D cells infected with a retroviral vector carrying AML1-MTG16 and unable of granulocyte differentiation and proliferation in response to the granulocyte colony stimulating factor (G-CSF), or from 32D cells infected with the cognate empty vector. From the analysis of differential gene expression alone (using as criteria a p value < 0.01 and an absolute fold change > 3), we were unable to conclude which of the 37 genes downregulated by AML1-MTG16 were, or not, direct AML1 targets. However, when we applied a bioinformatic approach to search for AML1-consensus sequences in the 10 Kb around the gene transcription start sites, we closed on 17 potential direct AML1 targets. By focusing on the most significantly downregulated genes, we found that both the AML1-consensus and the transcription start site chromatin regions were significantly marked by aberrant repressive histone tail changes. Further, the promoter of one of these genes, containing a CpG island, was aberrantly methylated.
Conclusion
This study shows that a leukemia-associated fusion protein can impose a distinct epigenetic repressive signature at specific sites in the genome. These findings strengthen the conclusion that leukemia-specific oncoproteins can induce non-random epigenetic changes.
Similar content being viewed by others
Background
Nuclear hormone receptors and transcription factors can regulate the transcription of their target genes by inducing chromatin changes. Paradigmatic are the retinoic acid receptor alpha (RARα) and the transcription factor core binding factor (CBF), which regulate in this way the transcription of target genes involved in hematopoietic processes [1, 2]. Differently from RARα, which epigenetically activates its targets by recruiting coactivator protein complexes with histone acetyl transferase (HAT) activity only when bound to retinoic acid, CBF can directly recruit HAT-containing complexes to activate its targets [3–6]. One of the two CBF subunits, CBFα or AML1, can bind target genes endowed with the AML1-consensus sequence TG(T/C)GGT via its N-terminal DNA-binding domain [7]. AML1, encoding a master hematopoietic transcription factor, is frequently affected by different chromosome translocations in leukemic cells [8]. Moreover, AML1 haploinsufficiency was found to be associated with familial platelet disorder, a condition predisposing to acute myeloid leukemia [9].
Two leukemia-associated chromosome translocations, the t(8;21)(q22;q22) and the t(16;21)(q24;q22), result in the fusion between the N-terminal region of AML1 and the C-terminal regions of two almost identical chromatin corepressors, MTG8 and MTG16, leading to the formation of AML1-MTG8 and AML1-MTG16, respectively [10–13]. Upon fusion with either MTG8 or MTG16, AML1 is converted from a transcriptional activator into a transcriptional repressor of AML1-targets. Specific MTG domains in the wild type, as well as in the MTG fusion proteins, can interact, directly or via other corepressors such as NCoR and Sin3A, with histone deacetylases (HDACs), thus creating a repressive chromatin state at AML1 target sites (reviewed in [14, 15]). Repression at these sites is further enhanced by the formation of oligomers between the fusion proteins and wild-type MTG proteins [16–18].
Myeloid cell differentiation systems, such as the 32D mouse myeloid cell line, ectopically expressing either AML1-MTG8 or AML1-MTG16, were used as models to simulate some of the effects of these fusion proteins in myelogenesis and leukemogenesis. Both fusion proteins, when exogenously expressed in the 32D background, were shown to affect granulocytic differentiation and produce distinct effects on cell proliferation [19–21]. In a preliminary study, we found that AML1-MTG16, when exogenously expressed in 32D cells, can induce aberrant myeloid phenotypes in association with repressive modifications at the chromatin of the Colony stimulating factor 1 receptor (Csf1r), an AML1-target gene encoding the macrophage colony stimulating factor receptor [19]. Based on this finding, we hypothesize that the comparative epigenetic analysis of the changes induced by different AML1-MTG fusion proteins in an identical cell context (e.g. the 32D context) might provide a lead to elucidating the differences observed in leukemic cells carrying either one of the two proteins [8]. The objective of this study was to demonstrate whether AML1-MTG16 induces epigenetic changes at AML1-target genes in the 32D myeloid cell genome. Only by coupling global gene expression array analysis with a bioinformatic genomic survey for the AML1-consensus sequence, we were able to close onto AML1-targets downregulated by AML1-MTG16. AML1-MTG16-induced transcriptional downregulation was marked by the acquisition of a distinct repressive chromatin signature.
Results
Global gene expression array analysis of AML1-MTG16-expressing cells
To study the molecular and biological consequences of AML1-MTG16 expression in a myeloid differentiation cell model, we previously developed, by infecting 32D mouse myeloblasts with retroviral particles carrying either the pLNCX2 vector containing the AML1-MTG16 cDNA or the cognate empty vector, stable independent clones expressing AML1-MTG16 (hereafter called A16 clones) and stable independent control clones (hereafter called "mock" clones), respectively (Figure 1A). Upon treatment with granulocyte colony stimulating factor (G-CSF), A16 clones do not undergo granulocytic differentiation and proliferate significantly less than mock clones (Figure 1B). Global gene expression analysis (setting the p-value at < 0.05 and the absolute fold change at > 1.5) of a prototypic A16 clone and a prototypic mock clone grown either with interleukin 3 (IL-3) or G-CSF for 16 h, was combined with bioinformatic analysis of the proteins encoded by all the differentially expressed genes with the Ingenuity software (see Methods). This analysis clearly revealed a network comprising proteins critical for platelet function in A16 cells (see Additional file 1). The identification of this protein network strongly supports the biological data, indicating the occurrence of functional AML1 haploinsufficiency in A16 cells [9].
Global gene expression analysis of AML1-MTG16-expressing cells. A. The 32D cell model, comprising clones expressing the AML1-MTG16 protein (A16 clones) and control clones ("mock" clones), which do not express the fusion protein. B. A16 clones, differently from mock clones, do not undergo granulocytic differentiation and display an impaired proliferation in the presence of G-CSF. C. Most of the genes whose expression is significantly affected in A16 cells were found previously implicated in biological processes.
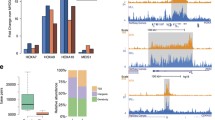
Further analysis of the gene expression data (setting the p-value at < 0.01 and the absolute fold change at > 3) enabled us to identify 138 differentially expressed genes, of which 66 differentially expressed genes in cells grown with IL-3, 67 differentially expressed genes in cells grown with G-CSF, and 5 differentially expressed genes in both cells grown with IL-3 and G-CSF (Figure 1C, left, and Table 1 and Table 2). According to the Ingenuity software, the differentially expressed genes in A16 cells were mostly implicated in tumorigenesis, cell proliferation, and hematopoiesis (Figure 1C, right). Since from this analysis alone we were unable to conclude whether, or not, these genes were AML1-MTG16 direct targets, we devised a bioinformatic approach aimed at identifying the AML1-consensus sequence in the 10 Kb region around the transcription start site of these genes.
Identification of genes containing the AML1-consensus sequence by bioinformatic analysis
Since the AML1-MTG proteins have a transcriptionally repressive function (reviewed in [14]), we focused our bioinformatic analysis on the 37 genes downregulated by AML1-MTG16 (see genes in bold in Table 1 and Table 2). Specifically, we searched the 10 Kb around the transcription start site of each gene for either the AML1-binding consensus sequence TG(T/C)GGT or, this sequence in reverse orientation, ACC(G/A)CA. With the MEME software (see Methods) we identified a conserved motif, hereafter called AML1-consensus motif (Figure 2A), encompassing the AML1-consensus sequence in seventeen out of the 37 genes (Figure 2B and Table 3). We focused on five of these genes, Fcer1a, Tcfec, Ptprcap, F2rl3, and Mgmt (Figure 2B, right), because they were among the most significantly downregulated genes. Fcer1a, Tcfec, Ptprcap, F2rl3, and Mgmt encode for known proteins. Specifically, Fcer1a is the Fc fragment of IgE and is involved in the immune response [22]; Tcfec is a transcription factor that induces, among other genes, the G-CSF receptor gene [23, 24]; Ptprcap is a transmembrane protein associated with CD45, a key regulator of lymphocytes activation [25]; F2rl3 is a member of G protein-coupled protease-activated receptors (PARs) of the coagulation factor II (thrombin) and plays an important role in platelet activation [26]; Mgmt is a DNA repair enzyme that is frequently lost in cancer due to epigenetic silencing [27]. Downregulation of these genes was confirmed by real time RT-PCR (Figure 2C).
AML1-MTG16-induced downregulation of putative AML1-targets. A. The AML1-consensus motif, containing the AML1-consensus sequence (framed), found by bioinformatic analysis of the genes significantly downregulated in A16 cells. The height of the columns associated with each nucleotide is proportional to the conservation level. The "logo" representation of the motif instead indicates in which proportion the single bases are present at each position. B. Seventeen out of the 37 downregulated genes are putative AML1-targets. The fold-changes of five of the most significantly downregulated genes are reported at right. C. Real time RT-PCR confirmed the significant (p < 0.01) downregulation of the five genes.
Fcer1a, Tcfec, Ptprcap, F2rl3, and Mgmt are direct AML1-MTG16 targets
Quantitative chromatin immunoprecipitation (ChIP) with an anti-AML1 specific antibody, but not with an anti-MTG16 antibody (data not shown), showed significant (p < 0.05) enrichment of the region encompassing the AML1-consensus motif (see bars in figure 3A, left) relative to an arbitrary control region without the AML1-consensus motif in the mock clone chromatin for all five genes, indicating endogenous AML1 binding at these regions (Figure 3B). ChIP with an anti-MTG16 antibody showed instead a significant enrichment of exogenous AML1-MTG16 in the same chromatin regions in the A16 clones (Figure 3B). The human homologues of these genes also contain an AML1-consensus sequence(s) in the 10Kb region surrounding the transcription start site, pointing to these five genes as novel, bona fide direct AML1-targets genes.
AML1-target gene validation. A. Relative position of the AML1-consensus motifs (left) and their sequence (right) in the five putative AML1-target genes that were analyzed by ChIP. B. Quantitative ChIP analysis with antibodies either against AML1 or MTG16 showing a significant (p < 0.05) enrichment of chromatin containing AML1-consensus motifs vs. chromatin containing a control region in AML1-MTG16-negative and AML1-MTG16-positive cells, respectively.
Repressive chromatin changes at AML1-MTG16-downregulated targets
We previously demonstrated that AML1-MTG16 interacts with both HDAC1 and HDAC3 [28]. Further, we found that AML1-MTG16 can induce downregulation marked by repressive histone hypoacetylation at the Csf1r chromatin [19]. Here we show that, in A16 cells, the chromatin associated with both the region containing the AML1-consensus motif and the region encompassing the transcription start site of Fcer1a, Tcfec, Ptprcap, F2rl3, and Mgmt (Figure 3A) displays a significant (p < 0.05) decrease of acetylated histone H4 (Ac-H4), and a significant (p < 0.05) increase of H3K9 tri-methylation (Tri-Met-H3-K9) (Figure 4A), supporting the acquisition of a repressive chromatin state [29–31].
Repressive epigenetic changes at the AML1-targets. A. ChIP with antibodies against either acetylated histone H4 or tri-methylated histone H3 Lysine 9 (tri-Met-H3-K9) followed by quantitative PCR with primers amplifying a region encompassing either the transcription start site (+1) or the AML1-consensus detected a different level of repressive histone changes in all five genes in A16 cells. B. In silico analysis identified a CpG island only in the Mgmt promoter. This CpG island is hypermethylated in A16 cells (bottom, right).
Repressive histone modifications are often associated with aberrant hypermethylation at CpG islands present in the 5' regulatory regions of many genes [32, 33] and references within). By using the CpG island searcher [34], a software for the identifying CpG islands, we could identify a CpG island only in the Mgmt promoter region [35] (Figure 4B). Bisulfite sequencing analysis of this region detected hypermethylation in AML1-MTG16-positive cells (Figure 4B).
The overall epigenetic analysis indicates that downregulation of AML1-targets by AML1-MTG16 can be achieved, even in the absence of DNA methylation, when there is a critical quantitative level of repressive histone changes.
Discussion
In this study we show the effectiveness of integrating global gene expression array analysis with a bioinformatic approach aimed at detecting AML1-consensus sequences for identifying novel putative direct AML1-targets downregulated by AML1-MTG16 in 32D cells. Downregulation of these genes is marked by a distinct repressive chromatin profile.
When we surveyed the 37 most significantly downregulated genes for the presence of the AML1-consensus motif(s) in the 10 Kb region encompassing the transcription start site, we closed on seventeen putative direct AML1-MTG16 targets. For five of these genes, Fcer1a, Tcfec, Ptprcap, F2rl3 and Mgmt, which were among the most significantly downregulated, we were able to demonstrate, using ChIP analysis, the binding of both AML1 and AML1-MTG16 to the gene regions containing the AML1-motifs. Thus, our two-tier approach, combining gene expression array analysis with bioinformatic survey for transcription factor-consensus sequences, seems to be a powerful strategy for identifying transcription factor targets, which would otherwise be missed when using conventional gene expression array analysis alone.
The chromatin of the five downregulated genes, Fcer1a, Tcfec, Ptprcap, F2rl3, and Mgmt, was marked not only by significant levels of histone H4 hypoacetylation, but also by significant levels of repressive histone H3-K9 trimethylation, suggesting that AML1-MTG16 might induce the recruitment of both histone deacetylases [28] and histone methyltransferases. Apparently, a critical quantity of repressive histone modifications, even in the absence of CpG methylation, might per se be sufficient to "lock in" a transcriptionally downregulated state. In the case of Mgmt, which has a CpG island, it is instead possible that the accumulation of histone repressive changes preceded CpG hypermethylation [[36], and references within].
It is noteworthy that all the genes for which we demonstrated AML1-MTG16-induced epigenetic downregulation encode for functions relevant to either hematopoiesis and/or leukemogenesis. We would like to underline that downregulation of two of the genes that we identified might be relevant to AML1-MTG16-induced leukemogenesis. One of these genes is Tcfec, whose human counterpart encodes a transcription factor that induces the granulocyte colony stimulating factor receptor G-CSFR [23, 24]. Remarkably, Tcfec downregulation in A16 cells is paralleled by a significant downregulation of G-csfr (data not shown), indicating that AML1-MTG16 might have triggered a coordinated cascade of transcriptional downregulation, as we observed in other differentiation model systems [37, 38]. The second gene is Mgmt, encoding the DNA repair enzyme O6-Methylguanine-DNA-methyltransferase, which is frequently silenced and hypermethylated in leukemia [39]. MGMT epigenetic silencing is thought to lead to random mutations in cancer [40]. A recent study has shown that expression of different acute myeloid leukemia fusion proteins, including AML1-MTG8, leads to downregulation of several DNA repair genes [41]. Thus, the induction of a "mutator phenotype" might be a common consequence of leukemia fusion protein expression.
A few global gene expression studies on cells expressing exogenous AML1-MTG8 have been recently described [42–44]. Given the use of different cell systems, it is difficult to compare the differentially expressed genes in AML1-MTG16-positive 32D cells with the differentially expressed genes reported for AML1-MTG8. Nevertheless, we could identify a few gene families (e.g. S100 Calcium-binding proteins) that are similarly affected by both AML1-MTG8 and AML1-MTG16 even in different cell contexts. Extending our study to the comparison of the epigenetic signatures imposed by either exogenous AML1-MTG16 or exogenous AML1-MTG8 in the very same cell context (e.g. 32D cells) might enable us to narrow down additional critical epigenetic signatures consequent to t(8;21) and t(16;21) translocations.
Conclusion
In this study, we show that AML1-MTG16, the leukemia fusion protein associated with the non-random chromosome translocation t(16;21)(q24;q22), can impose transcriptional downregulation marked by a distinct epigenetic signature at specific AML1-target sites in the genome. Thus, our findings further support the hypothesis that non-random genetic abnormalities can lead to non-random epigenetic changes in leukemia cells [19, 45].
Methods
Cell cultures
Stable clones obtained from mouse myeloid 32D cells infected either with pLNCX2-AML1-MTG16 (A16 clones) or the empty vector pLNCX2 (mock clones) were previously described [19]. Two prototypic A16 clones and two prototypic mock clones were used in this study. Cells were maintained in the presence of 10 ng/ml of murine IL-3 (BD Biosciences, San Jose, CA, USA) in RPMI 1640 medium supplemented with 10% fetal calf serum, 1% antibiotics (penicillin/streptomycin), adjusting the cell density to 2 × 105 cells/ml daily. To induce granulocyte differentiation, cells were washed in RPMI medium, and IL-3 was replaced with 10 ng/ml human G-CSF (Amgen, Thousand Oaks, CA, USA). Differentiation was microscopically evaluated on cytospin preparations stained with May-Grünwald-Giemsa.
RNA extraction and microarray hybridization
Total RNA was extracted with RNeasy mini kit (Qiagen, Hilden, Germany) and treated with DNase (Qiagen). Double stranded cDNA was generated from 5 μg RNA using Superscript ds cDNA synthesis kit (Invitrogen, Carlsbad, CA, USA) and T7-oligo(dT) primers. The cDNA was purified with GeneChip Sample Cleanup Module (Affymetrix, Santa Clara, CA, USA) and used to synthesize biotin-labeled cRNA with Enzo RNA transcript Labeling Kit (Enzo Life Science, Farmingdale, NY, USA). Purified cRNA was quantified by spectrophotometric methods and the concentration was adjusted in order to exclude the carryover of unlabeled RNA. 11 μg of cRNA were then fragmented in fragmentation buffer (Affymetrix) at 95°C for 35 minutes and hybridized for 16 h at 45°C onto MOE430A microarrays (Affymetrix). After washing and staining, the chips were scanned in a Hewett-Packard/Affymetrix scanner at 570 nm. For all the samples the 5'/3' ratios of Gapdh were 0.7 – 0.9. In comparative experiments the scaling factor, noise and presence calls were similar. Gene expression data represent the average of two independent experiments.
Microarray data analysis
The arrays were normalized by geometric mean intensity for each probe set and scaled using log2 transformation for further analysis. Comparison between the A16 and mock clones grown with either IL-3 or G-CSF was done using Spotfire Decision Site. This comparison generated a p-value from a t-test to statistically extract significant changes in mRNA expression levels between the groups. p-values < 0.05 were considered significant. The null hypothesis is that the samples between the groups are derived from the same population i.e. there is no significant differential expression. The t-test looks at the variance within the groups as well as between them. To be considered significantly differentially expressed the variance had to be greater between than within the groups to a level of p < 0.05. Ratios were generated by dividing the average of the unlogged control data by the average of the unlogged AML1-MTG16 data. Ratios were then portrayed as positive or negative fold change between A16 and mock. To confirm statistical significance of these ratios the differentially expressed genes had to satisfy an arbitrary cut-off ratio as well as having a p-value < 0.05 (see Results section). Analysis of the protein networks was performed by using Ingenuity Pathways Analysis (Ingenuity Systems, Redwood City, CA), software able to identify molecular networks based on known functional or physical interactions among the proteins encoded by the differentially expressed genes.
Search of AML1-consensus sequence in differentially expressed genes
The well-annotated genes differentially expressed in the A16 clone versus the mock clone either in the presence of IL-3 or G-CSF (p < 0.01 and absolute fold change >3) were searched for the AML1-consensus sequence "5'-TG(T/C)GGT-3"' in the 10 kb region surrounding the transcription initiation sites (from -5000 bp to +5000 bp) using an in-house built PERL script. A 400 bp sequence flanking the potential AML1-binding sites (200 bp on each side) was extracted and analyzed with MEME, which is a software package to discover motifs in groups of related DNA sequences [46], and with multiple sequence alignment to test whether additional conserved motifs in the surrounding regions could be identified and to assess the sequence conservation extending the potential AML1-binding sites.
Real-time RT-PCR
Total RNA was obtained using Trizol (Invitrogen), treated with DNase I (Ambion, Austin, TX, USA), retrotranscribed with SuperScript™ First-Strand Synthesis System (Invitrogen) and amplified by Real-time RT-PCR on an iCycler (Bio-Rad, Hercules, CA, USA) by using iQ SYBR Green Supermix (Bio-Rad) and primers specific for γ actin, F2rl3, Fcer1a, Ptprcap, Tcfec, and Mgmt (Table 4). Transcript levels of the genes of interest were quantitated by the Delta-delta Ct method, using the house keeping gene γ-actin for normalization. The amplification efficiency, evaluated from the sample slopes, was similar for all the samples analyzed in the same experiment. Two independent experiments were performed in triplicate using two mock clones and two A16 clones. Significance was determined by using the Student t-test.
Quantitative chromatin immunoprecipitation (ChIP)
ChIP was performed using reagents purchased from Upstate (Charlottesville, VA, USA) following the manufacturer's protocol. AML1 and AML1-MTG16 binding was assessed by ChIP with antibodies against either the AML1 C-terminus (Santa Cruz Biotechnology, Santa Cruz, CA, USA), or the MTG16 C-terminus [28], respectively. Histone hallmarks of repressive chromatin were assessed by ChIP with antibodies against acetyl-histone H4 (Upstate) and trimethyl-K9 at histone H3 (Upstate). Control ChIPs were performed without the respective antibodies. The immunoprecipitated DNA was amplified by real-time PCR with primers specific for regions encompassing the AML1-consensus, the transcription start site, or a control region (Table 4). The DNA relative enrichment was calculated by using the Delta-delta Ct method. The PCR signals obtained for each gene region were normalized to the PCR signal obtained from the input DNA (total chromatin fraction) and compared to a control region approximately 15 kb downstream of F2rl3 transcription start site. Two independent experiments were performed in triplicate, and significance was calculated by using the Student t-test.
Bisulfite sequencing
Genomic DNA was extracted with DNAzol (Invitrogen) according to the manufacturer's instructions. DNA was modified by sodium bisulfite treatment as previously described [47]. Mgmt CpG island was amplified by nested PCR by using the primers indicated in Table 4. The PCR fragments were subcloned into pGEM-T (Promega, San Luis Obispo, CA, USA) and 20 clones for each PCR fragment were sequenced.
References
Rosmarin AG, Yang Z, Resendes KK: Transcriptional regulation in myelopoiesis: Hematopoietic fate choice, myeloid differentiation, and leukemogenesis. Exp Hematol. 2005, 33: 131-143. 10.1016/j.exphem.2004.08.015.
Evans T: Regulation of hematopoiesis by retinoid signaling. Exp Hematol. 2005, 33: 1055-1061. 10.1016/j.exphem.2005.06.007.
Kitabayashi I, Yokoyama A, Shimizu K, Ohki M: Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. Embo J. 1998, 17: 2994-3004. 10.1093/emboj/17.11.2994.
Michaud J, Scott HS, Escher R: AML1 interconnected pathways of leukemogenesis. Cancer Invest. 2003, 21: 105-136. 10.1081/CNV-120018821.
Otto F, Lubbert M, Stock M: Upstream and downstream targets of RUNX proteins. J Cell Biochem. 2003, 89: 9-18. 10.1002/jcb.10491.
Yamagata T, Maki K, Mitani K: Runx1/AML1 in normal and abnormal hematopoiesis. Int J Hematol. 2005, 82: 1-8. 10.1532/IJH97.05075.
Meyers S, Downing JR, Hiebert SW: Identification of AML-1 and the (8;21) translocation protein (AML- 1/ETO) as sequence-specific DNA-binding proteins: the runt homology domain is required for DNA binding and protein-protein interactions. Mol Cell Biol. 1993, 13: 6336-6345.
Scandura JM, Boccuni P, Cammenga J, Nimer SD: Transcription factor fusions in acute leukemia: variations on a theme. Oncogene. 2002, 21: 3422-3444. 10.1038/sj.onc.1205315.
Song WJ, Sullivan MG, Legare RD, Hutchings S, Tan X, Kufrin D, Ratajczak J, Resende IC, Haworth C, Hock R, Loh M, Felix C, Roy DC, Busque L, Kurnit D, Willman C, Gewirtz AM, Speck NA, Bushweller JH, Li FP, Gardiner K, Poncz M, Maris JM, Gilliland DG: Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet. 1999, 23: 166-175. 10.1038/13793.
Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M: t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci U S A. 1991, 88: 10431-10434. 10.1073/pnas.88.23.10431.
Erickson P, Gao J, Chang KS, Look T, Whisenant E, Raimondi S, Lasher R, Trujillo J, Rowley J, Drabkin H: Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992, 80: 1825-1831.
Nisson PE, Watkins PC, Sacchi N: Transcriptionally active chimeric gene derived from the fusion of the AML1 gene and a novel gene on chromosome 8 in t(8;21) leukemic cells [published erratum appears in Cancer Genet Cytogenet 1993 Mar;66(1):81]. Cancer Genet Cytogenet. 1992, 63: 81-88. 10.1016/0165-4608(92)90384-K.
Gamou T, Kitamura E, Hosoda F, Shimizu K, Shinohara K, Hayashi Y, Nagase T, Yokoyama Y, Ohki M: The partner gene of AML1 in t(16;21) myeloid malignancies is a novel member of the MTG8(ETO) family. Blood. 1998, 91: 4028-4037.
Hiebert SW, Lutterbach B, Amann J: Role of co-repressors in transcriptional repression mediated by the t(8;21), t(16;21), t(12;21), and inv(16) fusion proteins. Curr Opin Hematol. 2001, 8: 197-200. 10.1097/00062752-200107000-00003.
Rossetti S, Hoogeveen AT, Sacchi N: The MTG proteins: chromatin repression players with a passion for networking. Genomics. 2004, 84: 1-9. 10.1016/j.ygeno.2004.02.011.
Minucci S, Maccarana M, Cioce M, De Luca P, Gelmetti V, Segalla S, Di Croce L, Giavara S, Matteucci C, Gobbi A, Bianchini A, Colombo E, Schiavoni I, Badaracco G, Hu X, Lazar MA, Landsberger N, Nervi C, Pelicci PG: Oligomerization of RAR and AML1 transcription factors as a novel mechanism of oncogenic activation. Mol Cell. 2000, 5: 811-820. 10.1016/S1097-2765(00)80321-4.
Zhang J, Hug BA, Huang EY, Chen CW, Gelmetti V, Maccarana M, Minucci S, Pelicci PG, Lazar MA: Oligomerization of ETO is obligatory for corepressor interaction. Mol Cell Biol. 2001, 21: 156-163. 10.1128/MCB.21.1.156-163.2001.
Liu Y, Cheney MD, Gaudet JJ, Chruszcz M, Lukasik SM, Sugiyama D, Lary J, Cole J, Dauter Z, Minor W, Speck NA, Bushweller JH: The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO's activity. Cancer Cell. 2006, 9: 249-260. 10.1016/j.ccr.2006.03.012.
Rossetti S, Van Unen L, Touw IP, Hoogeveen AT, Sacchi N: Myeloid maturation block by AML1-MTG16 is associated with Csf1r epigenetic downregulation. Oncogene. 2005, 24: 5325-5332. 10.1038/sj.onc.1208651.
Ahn MY, Huang G, Bae SC, Wee HJ, Kim WY, Ito Y: Negative regulation of granulocytic differentiation in the myeloid precursor cell line 32Dcl3 by ear-2, a mammalian homolog of Drosophila seven-up, and a chimeric leukemogenic gene, AML1/ETO. Proc Natl Acad Sci U S A. 1998, 95: 1812-1817. 10.1073/pnas.95.4.1812.
Kohzaki H, Ito K, Huang G, Wee HJ, Murakami Y, Ito Y: Block of granulocytic differentiation of 32Dcl3 cells by AML1/ETO(MTG8) but not by highly expressed Bcl-2. Oncogene. 1999, 18: 4055-4062. 10.1038/sj.onc.1202735.
Kinet JP: The high-affinity IgE receptor (Fc epsilon RI): from physiology to pathology. Annu Rev Immunol. 1999, 17: 931-972. 10.1146/annurev.immunol.17.1.931.
Rehli M, Sulzbacher S, Pape S, Ravasi T, Wells CA, Heinz S, Sollner L, El Chartouni C, Krause SW, Steingrimsson E, Hume DA, Andreesen R: Transcription factor Tfec contributes to the IL-4-inducible expression of a small group of genes in mouse macrophages including the granulocyte colony-stimulating factor receptor. J Immunol. 2005, 174: 7111-7122.
Rehli M, Lichanska A, Cassady AI, Ostrowski MC, Hume DA: TFEC is a macrophage-restricted member of the microphthalmia-TFE subfamily of basic helix-loop-helix leucine zipper transcription factors. J Immunol. 1999, 162: 1559-1565.
Altin JG, Sloan EK: The role of CD45 and CD45-associated molecules in T cell activation. Immunol Cell Biol. 1997, 75: 430-445.
Coughlin SR: Thrombin signalling and protease-activated receptors. Nature. 2000, 407: 258-264. 10.1038/35025229.
Gerson SL: MGMT: its role in cancer aetiology and cancer therapeutics. Nat Rev Cancer. 2004, 4: 296-307. 10.1038/nrc1319.
Hoogeveen AT, Rossetti S, Stoyanova V, Schonkeren J, Fenaroli A, Schiaffonati L, Van Unen L, Sacchi N: The transcriptional corepressor MTG16a contains a novel nucleolar targeting sequence deranged in t (16; 21)-positive myeloid malignancies. Oncogene. 2002, 21: 6703-6712. 10.1038/sj.onc.1205882.
Jenuwein T, Allis CD: Translating the histone code. Science. 2001, 293: 1074-1080. 10.1126/science.1063127.
Berger SL: Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002, 12: 142-148. 10.1016/S0959-437X(02)00279-4.
Peterson CL, Laniel MA: Histones and histone modifications. Curr Biol. 2004, 14: R546-51. 10.1016/j.cub.2004.07.007.
Fuks F: DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev. 2005, 15: 490-495. 10.1016/j.gde.2005.08.002.
Burgers WA, Fuks F, Kouzarides T: DNA methyltransferases get connected to chromatin. Trends Genet. 2002, 18: 275-277. 10.1016/S0168-9525(02)02667-7.
Takai D, Jones PA: The CpG island searcher: a new WWW resource. In Silico Biol. 2003, 3: 235-240.
Iwakuma T, Shiraishi A, Fukuhara M, Kawate H, Sekiguchi M: Organization and expression of the mouse gene for DNA repair methyltransferase. DNA Cell Biol. 1996, 15: 863-872.
Kouzarides T: Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002, 12: 198-209. 10.1016/S0959-437X(02)00287-3.
Bistulfi G, Pozzi S, Ren M, Rossetti S, Sacchi N: A repressive epigenetic domino effect confers susceptibility to breast epithelial cell transformation: implications for predicting breast cancer risk. Cancer Res. 2006, 66: 10308-10314. 10.1158/0008-5472.CAN-06-1052.
Pozzi S, Rossetti S, Bistulfi G, Sacchi N: RAR-mediated epigenetic control of the cytochrome P450 Cyp26a1 in embryocarcinoma cells. Oncogene. 2006, 25: 1400-1407. 10.1038/sj.onc.1209173.
Galm O, Wilop S, Luders C, Jost E, Gehbauer G, Herman JG, Osieka R: Clinical implications of aberrant DNA methylation patterns in acute myelogenous leukemia. Ann Hematol. 2005, 84 Suppl 13: 39-46. 10.1007/s00277-005-0005-0.
Esteller M, Herman JG: Generating mutations but providing chemosensitivity: the role of O6-methylguanine DNA methyltransferase in human cancer. Oncogene. 2004, 23: 1-8. 10.1038/sj.onc.1207316.
Alcalay M, Meani N, Gelmetti V, Fantozzi A, Fagioli M, Orleth A, Riganelli D, Sebastiani C, Cappelli E, Casciari C, Sciurpi MT, Mariano AR, Minardi SP, Luzi L, Muller H, Di Fiore PP, Frosina G, Pelicci PG: Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. J Clin Invest. 2003, 112: 1751-1761. 10.1172/JCI200317595.
Dunne J, Cullmann C, Ritter M, Soria NM, Drescher B, Debernardi S, Skoulakis S, Hartmann O, Krause M, Krauter J, Neubauer A, Young BD, Heidenreich O: siRNA-mediated AML1/MTG8 depletion affects differentiation and proliferation-associated gene expression in t(8;21)-positive cell lines and primary AML blasts. Oncogene. 2006, 25: 6067-6078. 10.1038/sj.onc.1209638.
Shimada H, Ichikawa H, Nakamura S, Katsu R, Iwasa M, Kitabayashi I, Ohki M: Analysis of genes under the downstream control of the t(8;21) fusion protein AML1-MTG8: overexpression of the TIS11b (ERF-1, cMG1) gene induces myeloid cell proliferation in response to G-CSF. Blood. 2000, 96: 655-663.
Shimada H, Ichikawa H, Ohki M: Potential involvement of the AML1-MTG8 fusion protein in the granulocytic maturation characteristic of the t(8;21) acute myelogenous leukemia revealed by microarray analysis. Leukemia. 2002, 16: 874-885. 10.1038/sj.leu.2402465.
Di Croce L, Raker VA, Corsaro M, Fazi F, Fanelli M, Faretta M, Fuks F, Lo Coco F, Kouzarides T, Nervi C, Minucci S, Pelicci PG: Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002, 295: 1079-1082. 10.1126/science.1065173.
Bailey TL, Elkan C: Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994, 2: 28-36.
Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB: Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996, 93: 9821-9826. 10.1073/pnas.93.18.9821.
Acknowledgements
We wish to thank Frank Staal, Justine Peeters, Violeta Stoyanova, Leontine van Unen (ErasmusMC, Rotterdam, The Netherlands), and Alan Hutson (Roswell Park Cancer Institute, Buffalo, NY) for technical support and critical discussions. This work was supported through Erasmus MC funds (ATH) and RPCI institutional funds (NS).
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
SR developed the 32D clones, designed and carried out the molecular genetics studies, participated in the microarray analysis, and prepared a draft of the manuscript. ATH contributed to the microarray analysis and critically reviewed the manuscript. PL performed the bioinformatic genome search of AML1-motifs. CS provided technical help for the real time RT-PCR and ChIP analyses. PV performed the microarray data analysis. NS conceived the hypothesis and critically reviewed the entire manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
12864_2006_751_MOESM1_ESM.pdf
Additional File 1: Evidence of functional AML1 haploinsufficiency in AML1-MTG16-expressing cells. This figure shows the Ingenuity Pathways Analysis of the global gene expression changes identified in AML1-MTG16-expressing cells. (PDF 101 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Rossetti, S., Hoogeveen, A.T., Liang, P. et al. A distinct epigenetic signature at targets of a leukemia protein. BMC Genomics 8, 38 (2007). https://doi.org/10.1186/1471-2164-8-38
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-8-38