Abstract
Background
Type I Interferons (IFNs) are well known cytokines which exert antiviral activity, antitumor activity and immunomodulatory effects. Single-nucleotide polymorphisms (SNP) and deletions in the gene coding for IFNA2 have been shown to influence the level of expression in vitro. The indel polymorphism -305_-300delAACTTT showed the strongest effect in vitro. To analyse the worldwide distribution of this polymorphism we analyzed five different populations (586 Vietnamese, 199 Central Africans, 265 Brazilians, 108 Kaingang and 98 Guarani). To investigate a possible association with susceptibility to infectious diseases we determined the polymorphism in malaria patients suffering either mild or severe malaria and in a cohort of hepatitis C virus infected individuals.
Results
We could detect the indel polymorphism in all populations analysed. There was no association with this polymorphism and the outcome of malaria but we found an increase of this indel polymorphism in hepatitis C virus positive individuals compared to healthy controls (p = 0.014).
Conclusion
Polymorphisms in genes involved in the interferon pathway have been implicated in the resistance or susceptibility against cerebral malaria and HBV. Here we show that an indel polymorphism, which mediates a disadvantageous effect in HBV patients, may also play a disadvantageous role in HCV infections stressing the importance of a fully functional interferon pathway.
Similar content being viewed by others
Background
Interferons (IFNs) constitute a family of secreted cytokines, which are expressed as an early response to various stimuli [1] in the first line of defense against viral infection [2]. The IFN family was originally recognized for its capacity to protect naïve cells against viral infection and IFN-α is known to play an important role in regulating and linking both the innate and the adaptive arms of immunity [3]. IFNs exhibit a broad diversity of biological functions, as represented by three major biological activities: antiviral activity, antitumor activity and immunomodulatory effects [4, 5]. The type I IFN family in humans comprise, in essence, 14 genes [6] coding for 13 interferons, and one IFN-beta and one IFN-omega [7]. They all lack introns and are clustered on the short arm of the chromosome 9 [8]. The IFN genes express different proteins that exhibit remarkably different activity profiles [9] and especially IFN-α2 has an important impact on the clinical field for the treatment of viral infections and different forms of cancer [6].
Recently it was shown that the genes IFNAR2 and IL10RB involved in binding interferons or interleukin 10-related cytokines, respectively were linked to persistence of hepatitis B virus in The Gambia [10]. This finding gave us a rational to look at other genes involved in the interferon pathway.
It has been shown that there is individual variation in the production of IFN-α [11] which could be due to mutations in the promoter of the IFNA2 gene. Song et al. (2006) showed that naturally occurring alterations in the promoter of this gene, especially the deletion -305_-300delAACTTT, reduced the transcription of this gene in vitro. This reduction could explain the individual differences in the interferon levels being one of the possible causes of susceptibility of hepatitis B [12].
In this paper we examined the level of genetic differentiation of the promoter of the IFNA2 gene by DNA sequencing and compared allele frequencies and genotype distributions in populations from Central Africa (Gabon), Brazil and Vietnam. One of these populations consisted of patients having malaria of different severity (Central Africans) [13]; the other was affected by hepatitis C virus (HCV) (Brazilians of Caucasian descent) [14]. With these patient cohorts we sought to test the hypothesis whether the indel polymorphism in the promoter of the IFNA2 gene is associated with disease outcome of other infectious diseases. Healthy Amerindians were included to investigate the frequency of this mutation and to detect a possible factor leading to an increased risk to contract infectious diseases [15].
Methods
Patients
1256 individuals were enrolled in this study at different sites: 586 healthy blood donors from the ethnic group of the Kinh at the Than Hung Dao Hospital in Hanoi as an extension of the study described in [12], 199 Central Africans at the Albert Schweitzer Hospital in Lambarènè [13] and 265 Brazilians of Caucasian origins at the Hospital de Clinicas in Curitiba [14], 108 Kaingang and 98 Guarani were recruited in their villages. Kaingang and Guarani are the two major Amerindian tribes living in Southern Brazil but despite living side by side for centuries the two groups differ in many aspects of culture and genetics; e. g. HLA type [16].
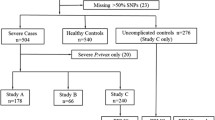
The Brazilians of European descent were either patients enrolled in a hepatitis C treatment study (n = 101) or healthy controls (n = 164) [14]. No patients had a history of alcohol or drug abuse. Patients were included into the study when HCV antibodies and HCV RNA was detected in serum; the presence of HBV and HIV antibodies was an exclusion criterion. From all patients biopsies were taken and liver histology was assessed blindly by two pathologists and staging of fibrosis.
Central Africans individuals were affected either by mild (n = 100) or severe malaria (n = 99) [13]. Mild malaria was characterised by a parasitaemia < 50,000 parasites/μl, haemoglobin > 80 g/L, glucose > 50 mg/dL, lactate < 3 mmol/L, leukocytes < 12/nL, platelets > 50/nL, no schizontaemia, and no signs of severe malaria such as cerebral malaria or other infections. The severely infected children presented with severe anaemia or hyperparasitaemia as the main complications of clinical disease. Appropriate treatment with sulfadoxine/pyrimethamine was given to mild cases. Severe cases were treated with intravenous quinine plus clindamycin.
Amerindians were healthy subjects from a human genetics study [17].
The genetic analysis was approved by the relevant ethic committees of the involved institutions: the Institutional Review Board of the Tran Hung Dao Hospital, Hanoi, Vietnam; the ethics committee of the Medical Faculty in Tübingen, Germany, the ethics committee of the Hospital de Clínicas in Curitiba, Brazil and the ethics committees of the International Foundation for the Albert Schweitzer Hospital in Lambaréné, Gabon.
DNA extraction
Blood from all the participants was collected in EDTA tubes and plasma was separated, aliquoted and stored at -20°C for later use. Genomic DNA was extracted from blood samples from using the QIAamp DNA blood kit (Qiagen, Hilden, Germany), as described in the user manual.
Polymerase chain reaction (PCR)
Genomic DNA was used as template for PCR. A single fragment of 480 bp from the IFNA2 promoter was amplified using the following primer sets: IFN-F: 5'-TTTCAAAAAAGTTGCTCTAAG-3', IFN-R: 5'-GTAGATGTTGCAGATGCTGCT-3' (Operon, Germany).
All amplifications were initiated with a denaturing step of 94°C for 5 min, followed by 42 cycles of 94°C for 30 sec, 45°C for 30 sec annealing time and 72°C for 1 minute elongation. The reaction was stopped after a final elongation step of 72°C for 10 min. A PTC-200 Peltier Termal Cycler (Bio-Rad, Munich, Germany) was used to amplify all the samples.
Genotyping
After purification, the respective PCR products were sequenced with the same primers described above using BigDye terminator chemistry on an ABI PRISM 3100 Genetic analyzer (Applied Biosystems, Foster City, USA). The sequences were analyzed for polymorphisms in the promoter of the IFNA2 gene after DNA alignment of the sequenced products using the Bioedit alignment program (North Carolina State University, USA). The quality of the sequencing was assessed by visual inspection with reference to the electropherograms.
Statistical analysis
IFNA2 allele frequencies and genotype distributions for each population as well as Hardy-Weinberg exact test were calculated using the Genepop version 3.4 http://genepop.curtin.edu.au/.
Differences in allele frequencies and genotype distributions between populations were calculated using Fisher's exact test using STATA http://www.stata.com/. P-value < 0.05 was considered to be significant.
Results
Allele frequency
Of the four mutations in the IFNA2 promoter only the deletion -305_-300delAACTTT was analyzed because it was shown to influence significantly the promoter activity in vitro [12]. The frequency of this deletion in Central Africans, Brazilians, Guarani, Kaingang and Vietnamese is presented in Table 1. Overall, 2512 alleles were analyzed, and 2246 out of 2512 (90%) had no deletion while 266 (10%) of the alleles carried the deletion.
Allele frequencies at this locus were compared for the five groups showing an irregular distribution in the different populations (P < 0.001). Therefore pairwise comparison of the allele frequencies was performed as shown in Table 2. The frequency of the deletion was significantly higher in Brazilians than in any other population except for the Guarani (P = 0.208). In Central Africans we find a significantly lower frequency of this variant allele compared with the rest of the populations except for Kaingang (P = 0.725).
Genotype distribution
The results of the observed genotype distribution in the five populations are shown in Table 3. Among the 1256 individuals enrolled in the study, 1010 (80.4%) had no deletion, 226 (18%) were heterozygous and 20 (1.6%) carried the deletion on both chromosomes (Table 3). The highest proportion of homozygous individuals carrying the deletion was found in the Brazilian population (5%). In contrast no homozygous individuals were found in the Guarani being the population with the highest number of heterozygous individuals. We performed pairwise comparison of the genotypes frequencies among the five groups (Table 4). Brazilians and Guarani did not significantly differ in genotype distribution whereas Central Africans were significantly different from any of the populations except for the Kaingang.
Hardy-Weinberg equilibrium
Expected genotype distributions were calculated using GenePop for the position which was polymorphic in the study populations (data not shown). The expected numbers did not significantly differ from the observed numbers in Central Africans, Guarani and Vietnamese, suggesting that these populations were in Hardy-Weinberg equilibrium. In contrast, exact tests indicated that expected and observed genotype distributions did significantly differ in Brazilians and Kaingang, indicating that these two groups are indeed in Hardy-Weinberg disequilibrium (Table 5).
Association of the deletion Δ-300/-305 with clinical aspects
To determine the association of the deletion -305_-300delAACTTT with different infectious diseases, we compared the frequency of IFNA2 with clinical manifestation of the diseases as well as with treatment outcome. No significant association of the mutation with severity of malaria (mild versus severe) were found in the Central African populations (allelic frequencies of the deletion in mild malaria cases 0.10 (90 individuals without deletion, 9 with the deletion on one chromosome) versus 0.12 in severe cases (89 individuals without deletion, 9 with the deletion on one chromosome, 1 individual with the deletion on both chromosomes, P = 1). There was a statistically significant association with the manifestation of HCV in Brazilians of Caucasian descent. We found among 101 HCV positive individuals 68 (67%) without deletion on any chromosome; 30 (30%) with a deletion on one chromosome and 3 (3%) with a deletion on both chromosomes. In the healthy control group we had in a total of 265 individuals 127 (77%) with no deletion on any chromosome; 27 (17%) with a deletion on one chromosome and 10 (6%) with a deletion on both chromosomes. The difference in the distribution is statistically significant (p = 0.03). When looking at the heterozygous individuals we find an odds ratio of 2 (CI = 1–4; p = 0.014) when we compare patients with healthy controls. The deletion was not found to be significantly associated with the treatment success of IFN-α with or without ribavirin in hepatitis C patients (data not shown).
Discussion
In this study we analyse a widely distributed chromosomal deletion in the promoter of the IFNA2 gene in five cohorts. It has been shown that the deletion -305_-300delAACTTT is associated with the clinical presentation of hepatitis B and a lower expression level in vitro [12]. Here we investigated a possible influence of this deletion in malaria and hepatitis C. There is evidence that IFN-α can induce NOS2 expression and NO production by human mononuclear cells in vitro and in viv o [18]. Also NO has been reported to have antiviral activity for a variety of viruses [19] and to play an important role in malaria [13, 20]. Vigário et al. have shown that recombinant human IFN-α inhibits cerebral malaria and reduces parasite burden in mice [21].
Additionally, IFN-α, either alone or in combination with ribavirin, is currently the standard treatment of patients with hepatitis C [22]. Therefore we speculated that the individual level of IFN-α depending on the promoter activity of the gene might affect the outcome of malaria or the therapy success in hepatitis C. In the latter event the intrinsic production would have supported the externally given IFN-α. We could not detect an influence of this polymorphism in Central Africans children with malaria. To detect a statistically significant distribution with frequencies in the malaria cohorts of 0.12 and 0.10 much larger cohorts would be needed.
Treatment success of HCV infection was not influenced by the presence of the polymorphism. We could, however, detect a difference in distribution of genotypes in HCV positive and healthy individuals. This finding confirms an earlier observation in hepatitis B patients stressing the importance of IFNA2 expression in these diseases.
The populations studied here diverged a long time ago and have undergone diverse selective pressure, explaining the genetic differences or similarities between groups. This indel polymorphism probably has an ancient origin and has entered the American continent 30.000 years ago [23] with the migration wave from Asia. Vietnamese and the two Amerindian groups, Kaingang and Guarani, therefore do not differ significantly in the genotype distribution. The Kaingang, unlike all other groups, do not show a significantly different genotype distribution to the Central Africans population, furthermore the Kaingang also diverge from the Hardy-Weinberg equilibrium. This finding is partly due to the traffic of slaves to South America during the 16th to the 19th century and a subsequent absorption of escaped African slaves by the Kaingang. By comparing blood groups and protein polymorphisms it was shown that Kaingang population had a higher proportion of non-Amerindian alleles than the Guaranis [24]. We found also a violation of the Hardy-Weinberg equilibrium in Brazilian, which is probably due to admixture in this area caused by the migration of individuals from different parts of Europe [25].
Conclusion
The polymorphism analysed in this work influences the course of hepatitis B and the prevalence of HCV. The treatment success in hepatitis C was not influenced by this indel polymorphism. Obviously, the externally given IFNα concentration is much higher than the intrinsic production of the patients. However, the fact that the indel polymorphism has is more frequently present in hepatitis B and C viruses argues for the importance of the IFNα pathway in both viral infections.
Human malaria was not influenced by the indel polymorphism; IFNα probably plays a role at the beginning of an infection but not at later time points when the parasite has manifested itself.
References
Doly J, Civas A, Navarro S, Uze G: Type I interferons: expression and signalization. Cell Mol Life Sci. 1998, 54: 1109-1121.
Sen GC: Viruses and interferons. Annu Rev Microbiol. 2001, 55: 255-281.
Dunn GP, Koebel CM, Schreiber RD: Interferons, immunity and cancer immunoediting. Nat Rev Immunol. 2006, 6: 836-848.
Takaoka A, Yanai H: Interferon signalling network in innate defence. Cell Microbiol. 2006, 8: 907-922.
Grandvaux N, tenOever BR, Servant MJ, Hiscott J: The interferon antiviral response: from viral invasion to evasion. Curr Opin Infect Dis. 2002, 15: 259-267.
Pestka S: The human interferon alpha species and receptors. Biopolymers. 2000, 55: 254-287.
Bonjardim CA: Interferons (IFNs) are key cytokines in both innate and adaptive antiviral immune responses – and viruses counteract IFN action. Microbes Infect. 2005, 7: 569-578.
Samuel CE: Antiviral actions of interferons. Clin Microbiol Rev. 2001, 14: 778-809.
Pestka S, Krause CD, Walter MR: Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004, 202: 8-32.
Frodsham AJ, Zhang L, Dumpis U, Taib NA, Best S, Durham A, Hennig BJ, Hellier S, Knapp S, Wright M, et al: Class II cytokine receptor gene cluster is a major locus for hepatitis B persistence. Proc Natl Acad Sci USA. 2006, 103: 9148-9153.
Hiscott J, Cantell K, Weissmann C: Differential expression of human interferon genes. Nucleic Acids Res. 1984, 12: 3727-3746.
Song LH, Toan NL, Xuan NT, Uhlemann AC, Boldt AB, Duy DN, Binh VQ, Kremsner PG, Kun JF: A promoter polymorphism in the interferon alpha-2 gene is associated with the clinical presentation of hepatitis B. Mutat Res. 2006, 601: 137-143.
Kun JFJ, Mordmüller B, Lell B, Lehman LG, Luckner D, Kremsner PG: Polymorphism in the promoter region of the inducible nitric oxide synthase gene and protection against malaria. Lancet. 1998, 351: 265-266.
Tena-Tomas C, Pedroso ML, Messias-Reason IJ, Kremsner PG, Kun JF: Polymorphisms in the IFNAR1 gene in patients with chronic hepatitis C: outcome of combined IFN-alpha therapy. Eur Cytokine Netw. 2007, 18: 136-141.
Tibayrenc M: Human genetic diversity and the epidemiology of parasitic and other transmissible diseases. Adv Parasitol. 2007, 64: 377-422.
Petzl-Erler ML, Luz R, Sotomaior VS: The HLA polymorphism of two distinctive South-American Indian tribes: the Kaingang and the Guarani. Tissue Antigens. 1993, 41: 227-237.
Weg-Remers S, Brenden M, Schwarz E, Witzel K, Schneider PM, Guerra LK, Rehfeldt IR, Lima MT, Hartmann D, Petzl-Erler ML, et al: Major histocompatibility complex (MHC) class III genetics in two Amerindian tribes from southern Brazil: the Kaingang and the Guarani. Hum Genet. 1997, 100: 548-556.
Sharara AI, Perkins DJ, Misukonis MA, Chan SU, Dominitz JA, Weinberg JB: Interferon (IFN)-α activation of human blood mononuclear cells in vitro and in vivo for nitric oxide synthase (NOS) type 2 mRNA and protein expression: possible relationship of induced NOS2 to the anti-hepatitis C effects of IFN-α in vivo. J Exp Med. 1997, 186: 1495-1502.
Benz D, Cadet P, Mantione K, Zhu W, Stefano G: Tonal nitric oxide and health: anti-bacterial and -viral actions and implications for HIV. Med Sci Monit. 2002, 8 (2): RA27-RA31.
Kun JFJ, Mordmüller BG, Perkins DJ, May J, Mercereau-Puijalon O, Alpers M, Weinberg JB, Kremsner PG: Nitric oxide synthase 2Lambaréné (G-954C), increased nitric oxide production and protection against malaria. J Infect Dis. 2001, 184: 330-336.
Vigario AM, Belnoue E, Gruner AC, Mauduit M, Kayibanda M, Deschemin JC, Marussig M, Snounou G, Mazier D, Gresser I, et al: Recombinant human IFN-alpha inhibits cerebral malaria and reduces parasite burden in mice. J Immunol. 2007, 178: 6416-6425.
Trepo C: Genotype and viral load as prognostic indicators in the treatment of hepatitis C. J Viral Hepat. 2000, 7: 250-257.
Bonatto SL, Salzano FM: Diversity and age of the four major mtDNA haplogroups, and their implications for the peopling of the New World. Am J Hum Genet. 1997, 61: 1413-1423.
Marrero AR, Silva-Junior WA, Bravi CM, Hutz MH, Petzl-Erler ML, Ruiz-Linares A, Salzano FM, Bortolini MC: Demographic and evolutionary trajectories of the Guarani and Kaingang natives of Brazil. Am J Phys Anthropol. 2007, 132: 301-310.
Price AL, Butler J, Patterson N, Capelli C, Pascali VL, Scarnicci F, Ruiz-Linares A, Groop L, Saetta AA, Korkolopoulou P, et al: Discerning the ancestry of European Americans in genetic association studies. PLoS Genet. 2008, 4: e236-
Acknowledgements
We thank all persons involved in the various studies for their cooperation. We are grateful to Andrea Weierich and Silvelia Grummes for excellent technical assistance in the DNA sequencing.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
CTT carried out the molecular genetic experiments, participated in the sequence alignment and drafted the manuscript. IMR supervised the HCV study. LHS participated in the sequence analysis. JT was involved in population genetics analysis. PGK designed the clinical study on malaria and supervised the performance. JFJK supervised the molecular work, was involved in the statistical analysis and finalized the manuscript.
All authors read and approved the final manuscript.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Tena-Tomás, C., de Messias-Reason, I., Song, L.H. et al. A globally occurring indel polymorphism in the promoter of the IFNA2 gene is not associated with severity of malaria but with the positivity rate of HCV. BMC Genet 9, 80 (2008). https://doi.org/10.1186/1471-2156-9-80
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2156-9-80