Abstract
Background
The salmon louse Lepeophtheirus salmonis is a parasitic copepod that infects salmonids in the Pacific and Atlantic oceans. Although considered as a single species, morphological and biological differences have been reported between lice from the two oceans. Likewise, studies based on nucleotide sequencing have demonstrated that sequence differences between Atlantic and Pacific L. salmonis are highly significant, albeit smaller than the divergence observed between congeneric copepod species.
Results
We demonstrated reproductive compatibility between L. salmonis from the two oceans and successfully established F2 hybrid strains using separate maternal lines from both the Pacific and Atlantic. The infection success for the F2 hybrid strains were similar to results typically observed for non hybrid lice strains in the rearing facility used. Lepeophtheirus salmonis COI and 16S sequences divergence between individuals from the Pacific and the Atlantic oceans was high compared to what may be expected within a copepod species and phylogenetic analysis showed that they consistently formed monophyletic clades representing their origin from the Pacific or Atlantic oceans.
Conclusions
Lepeophtheirus salmonis from the Pacific and Atlantic oceans are reproductively compatible at least until adults at the F2 hybrid stage, and should not be regarded as separate species based on reproductive segregation or sequence divergence levels. Reported biological and genetic differences in L. salmonis seen in conjunction with the reported genetic diversity commonly observed between and within species demonstrate that Atlantic and Pacific L. salmonis should be regarded as two subspecies: Lepeophtheirus salmonis salmonis and L. salmonis oncorhynchi subsp. nov.
Similar content being viewed by others
Background
The salmon louse (Lepeophtheirus salmonis Krøyer, 1838) is a marine ectoparasitic copepod found on salmonids in the northern hemisphere. This parasite causes large economic losses in commercial cage-based salmon aquaculture [1, 2], and has been causatively associated with declines in wild salmonid populations [3]. The latter of which is possibly linked with the fact that higher infestations of wild salmonids are typically observed in regions of intense commercial salmon farming [4–6]. Thus, L. salmonis represents both an economically and ecologically significant parasite in both the Pacific and Atlantic oceans which represent its natural distribution.
Lepeophtheirus salmonis displays a life cycle comprising eight stages: Two planktonic nauplius stages, an infective copepodid stage, two host-anchored chalimus stages and two motile preadult stages, before they ultimately molt into reproductive adults [7]. Thus, it displays considerable potential for dispersal, passively with ocean currents or while attached to its highly migratory hosts. This potential for dispersal has been confirmed by several recent genetics studies within the Atlantic [8–10] and Pacific [11] that have revealed panmixia or weak population genetic differentiation within each ocean basin. Once attached to its host, L. salmonis feeds on mucous and blood which causes physical damage, and may open wounds which can lead to osmoregulatory break-down and death in highly infected individuals [12, 13]. At more modest infection levels, L. salmonis causes the host stress and initiates a cascade of gene regulation associated with stress response functions [14, 15].
Several studies have investigated genetic differences between L. salmonis collected from the Pacific and Atlantic oceans. The first was based upon six microsatellite markers and revealed that 6% of the observed variation at these highly polymorphic loci was distributed among oceans [10]. A later analysis using the mtDNA gene cytochrome oxidase subunit 1 (COI) revealed 4.8-7.7% sequence divergence between Atlantic and Pacific L. salmonis[16]. By far the most comprehensive genetic comparison of L. salmonis from the Pacific and the Atlantic oceans has been conducted by Yazawa and colleagues [17]. Based upon the analysis of approximately 15 000 expressed sequence tags (ESTs), and sequence data covering the full mtDNA genome, these authors reported divergence between L. salmonis collected from these two oceans of 3.2% on average for nuclear genes, 7.1% for the entire mtDNA genome, 4.2% for ribosomal ribonucleic acid (rRNA) gene, and 6.1% for the COI gene. When the observed level of mtDNA divergence was compared with calibrated molecular clocks for copepods [18], it was concluded that the L. salmonis from the Pacific and Atlantic diverged approximately 2.5-11 million years ago. Additionally, reduced genetic variation has been observed among L. salmonis in the Pacific compared to the Atlantic leading Yazawa et al. to suggest that the species first established in the Atlantic and then in the Pacific following a limited introduction [17] when the Bering Strait opened approximately 5 million years ago [19].
Based upon the genetic diversity measures above, it has been proposed that L. salmonis from the Pacific and Atlantic may be regarded as separate forms of the same species [17] or even separate species [16]. Potentially in support of these suggestions is the fact that biological differences have also been identified between L. salmonis from these two oceans. These include reported differences in size [20, 21], time of development [22, 23] and possible differences in salinity tolerance [24, 25]. Furthermore L. salmonis from the Pacific have been observed frequently naturally occurring [26, 27] on three-spined sticklebacks (Gasterosteus aculeatus) while similar observations of common presence on non salmonid hosts have not been reported for L. salmonis in the Atlantic (but see [28, 29]).
Lepeophtheirus salmonis have been investigated in laboratory studies for approximately three decades, and recently, extensive breeding facilities and rearing protocols have been refined making controlled crossing experiments possible [30]. Here, we report a study where we established two F2 generation hybrid strains of L. salmonis with genetic contribution from Pacific and Atlantic L. salmonis and address the questions: are they reproductively compatible and should they be regarded as one species?
Methods
Lepeophtheirus salmonisculturing and hybrid production
The rearing experiments described here were conducted at the Institute of Marine Research’s experimental facility in Bergen, Norway. This laboratory has a quarantine facility with permit issued by the Norwegian Food Safety Authority (NFSA) to conduct experiments on marine pathogens and includes chlorination of all waste-water to ensure that no pathogens are released to the natural environment. Lice were cultured using well-established rearing protocols for L. salmonis[30]. During the experiments Atlantic salmon were kept in running seawater and fed commercial fish feed ad libitum. This experiment was conducted in accordance with Norwegian legislation for use of animals in research, and was approved by the Norwegian Animal Research Authority (research permit nr. 2009/186329).
Lice were cultured by infecting Atlantic salmon (Salmo salar) with copepodids from Atlantic and Pacific strains of L. salmonis. The Atlantic strain (LsAtl) was established by mixing approximately equal numbers of copepodids from the previously described LsGulen and LsOslofjord strains [30]. The Pacific strain (LsPac) was established from copepodids derived from adult female L. salmonis collected at a commercial salmon farm located close to the town of Campbell River (British Columbia, Canada). The Pacific lice were transported to Norway in thermal flasks containing full strength salinity seawater. A permit to import these lice was obtained from the NFSA (Additional file 1). Unfertilized L. salmonis from the LsPac F1 and the LsAtl F2 generations were sorted according to sex at the pre-adult II stage and used immediately (LsAtl) or kept separate (LsPac) on previously uninfected fish until used in the crossing experiment.
To obtain hybrid strains we crossed LsPac females with LsAtl males and LsAtl females and LsPac males using a similar tank rearing system to that described by Hamre and Nilsen [31]. Briefly, each cross consisted of two (in one instance one) females and one male which were placed on a single uninfected Atlantic salmon that was isolated in its own tank. This allowed definite control of the parent material and offspring generation. As the unfertilized females were in different stages of development (pre-adult II and adults), the time required to obtain fertilized egg strings from the two hybrid strains were dissimilar. After 15 (LsPac females and LsAtl males) and 35 (LsAtl females and LsPac males) days, females bearing egg strings, and the males used to fertilize them in the single fish tanks, were harvested. These sampled adults were stored in individual tubes containing 100% ethanol, and were indexed so that both parents and egg strings could be subsequently matched. The egg strings from the sampled females were incubated in separate incubators after removal of approximately 3-5mm of both egg strings which was stored in 100% ethanol. The parents and the 3-5mm section of egg strings were genotyped (described below) to validate the crosses by parentage assignment before the resulting copepodids were used to infect previously uninfected Atlantic salmon to produce the LsAtlPac (copepodids from LsAtl females and LsPac males) and LsPacAtl (copepodids from LsPac females and LsAtl males) F1 hybrid strains. The F1 hybrid strains were reared separately in replicate fish tanks (Figure 1).
The F1 generation hybrids were allowed to develop, fertilize and reproduce naturally on Atlantic salmon in their respective tanks. Approximately 3 months after infection with F1 hybrid copepodids, eggstrings from F1 hybrid females that had been fertilized by F1 hybrid males were harvested and incubated in separate containers for LsAtlPac and LsPacAtl. Copepodids arising from these egg strings were thereafter used to infect groups of previously uninfected Atlantic salmon in order to establish the LsAtlPac F2 and LsPacAtl F2 hybrid generation in two replicate tanks for each strain. The resulting F2 hybrid generations were allowed to develop until the pre-adult stage to allow sex determination before the experiments were terminated. Numbers of adults contributing to each generation were recorded. In addition, the numbers of copepodids that were used to propagate the F2 generation was estimated by counting an aliquot, and the numbers of pre-adults harvested from the F2 generation upon termination of the experiment, were determined by counting and sex determining all the lice present.
Genotype validation of F0 parents
In order to verify contributions of the individual pairs of parents to each set of hybrid egg strings for the F1 generation, both parents and an approximately 3-5mm section of each egg string was genotyped with microsatellite markers. DNA was extracted using a Qiagen DNeasy®96 Blood & Tissue Kit, and 16 microsatellite markers that have been recently used in a population genetic study of L. salmonis in the Atlantic ocean [8] were genotyped. These markers were amplified in three multiplex reactions. Multiplex 1; LsalSTA1, LsalSTA2, LsalSTA4, LsalSTA5[10], LsNUIG14 adapted by [10], multiplex 2; Lsal103EUVC, Lsal109EUVC, Lsal110EUVC, Lsal111EUVC[11], LsNUIG09[32], multiplex 3; Lsal104EUVC, Lsal105EUVC, Lsal106EUVC, Lsal108EUVC,[11], LsalSTA3[10], LsNUIG35B[33]. Amplification conditions are given in Additional file 2. PCR fragments were separated on an ABI 3730XL sequencer and sized relative to the Applied Biosystem GeneScan™–500LIZ™ size standard. Alleles were scored using automatic binning implemented in the Genmapper software (V4.0). Allele profiles were manually inspected to validate that egg strings contained a maximum of 4 alleles per locus (i.e., two alleles from each parent if both were heterozygotes for the locus), and that all alleles matched with both parents where available.
Comparing the Pacific and Atlantic 16S and COI sequences
To facilitate sequence comparison of 16S rRNA (16S) and cytochrome oxidase subunit I (COI) between the Pacific and Atlantic lice that were hybridized, the sequences were amplified and sequenced as described in Additional file 3. The obtained 16S sequences from the Pacific holotype specimen [GenBank: KF278676] and a representative female from the Atlantic founder generation for the F1 hybrids [GenBank:KF278677] were aligned with previously sequenced L. salmonis 16S sequences [9, 17] from the Pacific [GenBank:EU288264-EU288330] and the Atlantic [GenBank:AY602770-AY602949]. The obtained COI sequences from the Pacific holotype specimen [GenBank:KF278676] and a representative female from the Atlantic founder generation for the F1 hybrids [GenBank:KF278677] were aligned with previously sequenced L. salmonis COI sequences [9, 17] from the Pacific [GenBank:EU288201-EU288263] and the Atlantic [GenBank:AY602587-AY602766]. All sequences were aligned using CLC molecular workbench (6.8.2) at default settings and trimmed to the same length; 16S positions 84-879 (796 bp) and COI positions 121-1420 (1300 bp). All positions containing gaps and missing data were eliminated. Maximum likelihood phylogeny trees were constructed by MEGA5 [34] for each alignment using the Tamura-Nei model [35] and uniform rates for nucleotide substitution. Initial tree(s) for the heuristic search were obtained by applying the Neighbor-Joining method to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach. Support for each of the two consensus trees were calculated by bootstrapping 1000 times.
Results
The two founder strains (LsAtl and LsPac) were successfully established and subsequently used to produce the two F1 hybrid strains. These hybrid strains were thereafter propagated to produce two F2 hybrid strains. The number of adult females used to produce the copepodids founding each generation and the numbers of copepodids and resulting offspring for the hybrid strains are given in Tables 1 and 2. The low number of founding parents for the F1 hybrid strains (LsPacAtl and LsAtlPac) were caused by low fertilization success in the single fish tanks (each of which contained 2 females and a single male). In addition, some of the DNA samples taken from the egg strings from successfully fertilized females did not yield sufficient DNA to permit genotype validation of maternal and paternal contribution. These egg strings were not used. To establish the LsPacAtl F1 strain, the genetic contribution from both parents was validated with the allele profile of the egg strings. For the LsAtl F1 strain, the maternal origin was confirmed for all three pairs of egg strings used to establish the strain, but the paternal origin was only confirmed for one of these pairs. The last two pairs of egg strings shared the same father (i.e., were reared in the same single fish tank), but the father had been lost before sampling. Paternal contribution was however identical between these half-sibling families.
To tentatively estimate the fitness of the F2 hybrid strains, the numbers of copepodids used for infection of each replicate tank, and the numbers of pre-adult males and females harvested from the salmon, were recorded (Table 2). The infection success varied between 17 and 30%. Males were slightly overrepresented in all replicate tanks for both strains.
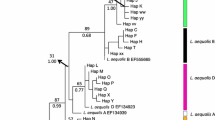
The phylogenetic analysis showed that the Atlantic and Pacific sequences were found in separate clusters with 100% bootstrapping support for both 16S rRNA and cytochrome oxidase subunit I (Figures 2 and 3). The phylogenetic relationship within each ocean based on 16S and COI sequences has previously been the topic of detailed studies [9, 17] and will not be discussed here.
Molecular Phylogeny of cytochrome oxidase subunit I (COI). The evolutionary history of 245 nucleotide partial COI sequences was inferred by using the Maximum Likelihood method. The tree with the highest log likelihood (-11199) is shown. The analysis reveals two distinct phylogenetic clades with 100% bootstrap support corresponding to the Atlantic and Pacific samples respectively. The Pacific and Atlantic entries from the present study are highlighted in yellow and red. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The Ls COI Pac sequence [GenBank:KF278676] was derived from the holotype speciemen (ZMUB91335). The Ls COI ATL sequence [GenBank:KF278677] was derived from a random female from the LsAtl strain.
Molecular Phylogeny of 16S ribosomal RNA. The evolutionary history of 249 nucleotide partial 16S rRNA sequences was inferred by using the Maximum Likelihood method. The tree with the highest log likelihood (-2387) is shown. The analysis reveals two distinct clades with 100% bootstrap support corresponding to the Atlantic and Pacific samples respectively. The Pacific and Atlantic entries from the present study are highlighted in yellow and red. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The Ls 16S Pac sequence [GenBank:KF278676] was derived from the holotype speciemen (ZMUB91335). The Ls 16S ATL sequence [GenBank:KF278677] was derived from a random female from the LsAtl strain.
Nomenclatural acts
The electronic edition of this article conforms to the requirements of the amended International Code of Zoological Nomenclature. The ZooBank LSIDs (Life Science Identifiers) can be resolved and the associated information viewed through any standard web browser by appending the LSID to the prefix “http://zoobank.org/”. The LSID for this publication is: urn:lsid:zoobank.org:pub:8CABA147-FB3D-4DFA-AF37-7FFB74AFB454.
Discussion
While the species concept and a single unequivocal definition of species remains a topic of discussion, most regard reproductive incompatibility between two groups of individuals as irrefutable evidence of separate species. Thus, reproductive incompatibility between L. salmonis from the Pacific and Atlantic would support the case for separating them into separate species as has been previously suggested based upon their COI genetic diversities [16]. In the present study, we successfully produced F1 and F2 hybrid L. salmonis strains using maternal lines from both the Pacific and the Atlantic. Previous studies of separate conspecific populations of copepods have shown that although defined as single species, the populations may or may not be reproductively compatible [36–38]. In studies of the intertidal copepod Tigriopus californicus it has been shown that “Dobzhansky-Muller incompatibilities” between strains, i.e. deleterious introductions of genetic variants into new genetic backgrounds, may result in reduced fitness in the F2 hybrids [39, 40]. Although the present study was primarily designed to qualitatively investigate the reproductive compatibility of Atlantic and Pacific L. salmonis, egg string hatchability and survival between infection and the pre-adult stage was quantified for the two hybrid strains in the F2 generation. The observed values (Table 1) fall within the range of values for these parameters in this salmon louse rearing facility [30, 41–43]. The infectivity and survival of Pacific L. salmonis reared in our facilities was not accurately quantified but did not appear to deviate from levels observed for Atlantic strains. Therefore, there are no data suggesting loss of fitness due to hybridization between Pacific and Atlantic L. salmonis up to and including the F2 hybrid generation. The genetic mtCOI divergence between T. californicus populations varies from 0.2% - 23% and the level of fitness reduction is correlated with the genetic divergence [44]. The genetic divergence between Atlantic and Pacific L. salmonis varies between 4.8 and 7.1% [16] and is thus limited when compared to the differences reported for T. californicus. This may suggest that outbreeding depression in hybrids between L. salmonis from the Pacific and Atlantic oceans may be expected to be limited if present, and could have gone undetected in the present study. It is therefore concluded that Pacific-Atlantic hybrid fitness should be accurately quantified in future studies, but that L. salmonis from the Pacific and Atlantic are reproductively compatible at least until the F2 hybrid generation. Therefore, our results do not support separating Atlantic and Pacific L. salmonis into separate species.
The COI gene has been sequenced for a large number of species and is currently being used for species identifications via DNA barcoding [45, 46]. Looking at congeneric species (species within the same genus) across a wide range of taxa, 98% of species-pairs display COI divergence of 2% or more, and the average divergence across all taxa is 11.3% [46]. Within Crustacea, the average diversity among congeneric species has been estimated to 15.4% [46]. Looking specifically at copepods, congeneric COI sequence diversity was estimated between 13-22% [47]. The calculated divergence for COI between L. salmonis from the Pacific and Atlantic (4.8-7.7%) [16, 17] is lower than what is typical for congeneric copepod species, but at the upper boundary of the reported variation of 1.3-7.9% within a crustacean species (i.e. between individuals of the same species) [48]. However, the congeneric calanoids Calanus glacialis and C. finmarchicus, with a reported COI sequence divergence of 20% [47], produce hybrids capable of successful reproduction [49]. Therefore, reproductive compatibility among Pacific and Atlantic L. salmonis may not be unexpected. Taken together, the reported sequence divergence of L. salmonis in the Pacific and the Atlantic oceans, which is well below the variations reported between congeneric copepods, and the evidence of reproductive compatibility with no apparent loss of fitness presented here, suggests that L. salmonis from the Pacific and Atlantic do not represent separate species.
While the genetic diversity within COI and 16S between L. salmonis from the Pacific and Atlantic is below that expected between species of copepods, it remains highly significant as illustrated by the phylogenetic analysis. This demonstrates minimal or non-existent genetic exchange between these allopatric L. salmonis components on a contemporary time-scale. This is in contrast to the very high level of gene-flow and lack of population differentiation reported among L. salmonis sampled from geographically distinct regions within each ocean [8, 10, 11, 17]. Stable barcoding gene sequence divergences between morphologically and geographically defined subspecies, below the divergence level expected between congeneric species, has previously been used to confirm subspecies validity [50, 51]. Lepeophtheirus salmonis from the Pacific and Atlantic exhibit morphological [20, 21] and apparent biological differences [24, 25, 27] in addition to considerable genetic sequence divergence [17]. Despite these differences, Atlantic and Pacific L. salmonis are reproductively compatible and exhibit sequence divergence below the level typically observed between congeneric copepods and at the extreme upper boundary of the range found among conspecific crustaceans [48]. Therefore, we suggest that Atlantic and Pacific L. salmonis should be regarded as two subspecies: Lepeophtheirus salmonis salmonis and Lepeophtheirus salmonis oncorhynchi subsp. nov. urn:lsid:zoobank.org:pub:8CABA147-FB3D-4DFA-AF37-7FFB74AFB454.
Due to the interaction of salmon lice with farmed and wild salmonids in both the Pacific and Atlantic, L. salmonis represents both an ecologically and economically significant parasite. In accordance with this is the growing volume of scientific studies investigating this parasite. The differences between L. salmonis in the two oceans make it crucial to correctly assign scientific results to the correct component of the species, which is presently taxonomically impossible. The proposed division is therefore well reasoned and will facilitate appropriate taxonomic indication of origin in the future.
Taxonomic summary of Lepeophtheirus salmonis oncorhynchisubsp. nov
Species: Lepeophtheirus salmonis oncorhynchi subsp. nov.
Etymology: ‘oncorhynchi’ reflecting the evolutionary association with the salmonid genus Oncorhynchus in the Pacific Ocean
Descriptions: Johnson and Albright [20, 22]
Holotype sequences: Lepeophtheirus salmonis oncorhynchi COI and 16S [GenBank:KF278676]
Museum specimens: Adult female ZMUB91335 (holotype, ethanol), adult male ZMUB91339 (allotype, karnovsky), paratype series, 3 specimens (ZMUB91336 – ZMUB91338, ethanol).
Conclusions
Lepeophtheirus salmonis from the Pacific and the Atlantic oceans are reproductively compatible at least until the F2 hybrid generation, and evaluation of F2 hybrids does not indicate reduced fitness among the hybrids. The genetic divergences of COI and 16S between the Atlantic and the Pacific populations are below what one expects to find between crustacean species but high compared to what one may expect to find within a species. Taken together these results indicate that Pacific and Atlantic L. salmonis should not be regarded as separate species. However, morphological, genetic and indicated ecological differences are significant and salmon louse from the Pacific and the Atlantic oceans should therefore be regarded as two subspecies: Lepeophtheirus salmonis salmonis and Lepeophtheirus salmonis oncorhynchi subsp. nov. This is not only scientifically appropriate, but is of practical significance as this will facilitate taxonomic assignment of results obtained with either subspecies, something that has not been possible hitherto.
References
Johnson SC, Treasurer JW, Bravo S, Nagasawa K, Kabata Z: A review of the impact of parasitic copepods on marine aquaculture. Zool Stud. 2004, 43: 229-243.
Costello MJ: The global economic cost of sea lice to the salmonid farming industry. J Fish Dis. 2009, 32: 115-118. 10.1111/j.1365-2761.2008.01011.x.
Krkosek M, Lewis MA, Volpe JP: Transmission dynamics of parasitic sea lice from farm to wild salmon. Proc Roy Soc B Biol Sci. 2005, 272: 689-696. 10.1098/rspb.2004.3027.
Tully O, Gargan P, Poole WR, Whelan KF: Spatial and temporal variation in the infestation of sea trout (Salmo trutta L.) by the caligid copepod Lepeophtheirus salmonis (Kroyer) in relation to sources of infection in Ireland. Parasitology. 1999, 119: 41-51. 10.1017/S003118209900445X.
Bjorn PA, Finstad B: Salmon Lice, Lepeophtheirus Salmonis (Kroyer), Infestation in Sympatric Populations of Arctic Char, Salvelinus Alpinus (L.), And Sea Trout, Salmo Trutta (L.), In Areas Near and Distant From Salmon Farms. Ices J Mar Sci. 2002, 59: 131-139. 10.1006/jmsc.2001.1143.
Morton A, Routledge R, Peet C, Ladwig A: Sea lice (Lepeophtheirus salmonis) infection rates on juvenile pink (Oncorhynchus gorbuscha) and chum (Oncorhynchus keta) salmon in the nearshore marine environment of British Columbia, Canada. Can J Fish Aquat Sci. 2004, 61: 147-157. 10.1139/f04-016.
Hamre LA, Eichner C, Caipang CM, Dalvin S, Bron JE, Nilsen F, Boxshall G, Skern-Mauritzen R: The Salmon Louse Lepeophtheirus salmonis (Copepoda: Caligidae) Life Cycle Has Only Two Chalimus Stages. PLoS ONE. 2013, 8 (9): e73539-10.1371/journal.pone.0073539.
Glover KA, Stolen AB, Messmer A, Koop BF, Torrissen O, Nilsen F: Population genetic structure of the parasitic copepod Lepeophtheirus salmonis throughout the Atlantic. Mar Ecol Prog Ser. 2011, 427: 161-172.
Tjensvoll K, Glover KA, Nylund A: Sequence variation in four mitochondrial genes of the salmon louse Lepeophtheirus salmonis. Dis Aquat Organ. 2006, 68: 251-259.
Todd CD, Walker AM, Ritchie MG, Graves JA, Walker AF: Population genetic differentiation of sea lice (Lepeophtheirus salmonis) parasitic on Atlantic and Pacific salmonids: analyses of microsatellite DNA variation among wild and farmed hosts. Can J Fish Aquat Sci. 2004, 61: 1176-1190. 10.1139/f04-069.
Messmer AM, Rondeau EB, Jantzen SG, Lubieniecki KP, Davidson WS, Koop BF: Assessment of population structure in Pacific Lepeophtheirus salmonis (Kroyer) using single nucleotide polymorphism and microsatellite genetic markers. Aquaculture. 2011, 320: 183-192. 10.1016/j.aquaculture.2010.09.033.
Dawson LHJ, Pike AW, Houlihan DF, McVicar AH: Effects of salmon lice Lepeophtheirus salmonis on sea trout Salmo trutta at different times after seawater transfer. Dis Aquat Organ. 1998, 33: 179-186.
Bjorn PA, Finstad B: The Development of Salmon Lice (Lepeophtheirus Salmonis) on Artificially Infected Post Smolts of Sea Trout (Salmo Trutta). Can J Zool. 1998, 76: 970-977. 10.1139/cjz-76-5-970.
Skugor S, Glover KA, Nilsen F, Krasnov A: Local and systemic gene expression responses of Atlantic salmon (Salmo salar L.) to infection with the salmon louse (Lepeophtheirus salmonis). BMC Genomics. 2008, 9: 498-10.1186/1471-2164-9-498.
Krasnov A, Skugor S, Todorcevic M, Glover KA, Nilsen F: Gene expression in Atlantic salmon skin in response to infection with the parasitic copepod Lepeophtheirus salmonis, cortisol implant, and their combination. BMC Genomics. 2012, 13: 130-10.1186/1471-2164-13-130.
Boulding EG, deWaard JR, Ang KP, Hebert PN: Population genetic structure of the salmon louse, Lepeophtheirus salmonis (Kroyer) on wild and farmed salmonids around the Pacific coast of Canada. Aquac Res. 2009, 40: 973-979. 10.1111/j.1365-2109.2008.02159.x.
Yazawa R, Yasuike M, Leong J, von Schalburg KR, Cooper GA, Beetz-Sargent M, Robb A, Davidson WS, Jones SRM, Koop BF: EST and Mitochondrial DNA Sequences Support a Distinct Pacific Form of Salmon Louse, Lepeophtheirus salmonis. Mar Biotechnol. 2008, 10: 741-749. 10.1007/s10126-008-9112-y.
Rocha-Olivares A, Fleeger JW, Foltz DW: Decoupling of molecular and morphological evolution in deep lineages of a meiobenthic harpacticoid copepod. Mol Biol Evol. 2001, 18: 1088-1102. 10.1093/oxfordjournals.molbev.a003880.
Marincovich L, Gladenkov AY: Evidence for an early opening of the Bering Strait. Nature. 1999, 397: 149-151. 10.1038/16446.
Johnson SC, Albright LJ: The developmental stages of Lepeophtheirus salmonis (Krøyer, 1837) (Copepoda, Caligidae). Can J Zool. 1991, 69: 929-950. 10.1139/z91-138.
Schram TA: Supplementary descriptions of the developmental stages of Lepeophtheirus salmonis (Krøyer, 1837) (Copepoda: Caligidae). Pathogens of Wild and Farmed Fish: Sea Lice. Edited by: Boxshall GA, Defaye D. 1993, Chichester, UK: Ellis Horwood, 30-47.
Johnson SC, Albright LJ: Development, Growth, and Survival of Lepeophtheirus-Salmonis (Copepoda, Caligidae) under Laboratory Conditions. J Mar Biol Assoc UK. 1991, 71: 425-436. 10.1017/S0025315400051687.
Heuch PA, Nordhagen JR, Schram TA: Egg Production in the Salmon Louse [Lepeophtheirus Salmonis (Kroyer)] in Relation to Origin and Water Temperature. Aquac Res. 2000, 31: 805-814. 10.1046/j.1365-2109.2000.00512.x.
Johnson SC, Albright LJ: Comparative Susceptibility and Histopathology of the Response of Naive Atlantic, Chinook and Coho Salmon to Experimental-Infection with Lepeophtheirus-Salmonis (Copepoda, Caligidae). Dis Aquat Organ. 1992, 14: 179-193.
Bricknell IR, Dalesman SJ, O’Shea B, Pert CC, Luntz AJM: Effect of environmental salinity on sea lice Lepeophtheirus salmonis settlement success. Dis Aquat Organ. 2006, 71: 201-212.
Jones S, Kim E, Dawe S: Experimental infections with Lepeophtheirus salmonis (Kroyer) on threespine sticklebacks, Gasterosteus aculeatus L., and juvenile Pacific salmon, Oncorhynchus spp. J Fish Dis. 2006, 29: 489-495. 10.1111/j.1365-2761.2006.00742.x.
Jones SRM, Prosperi-Porta G, Kim E, Callow P, Hargreaves NB: The occurrence of Lepeophtheirus salmonis and Caligus clemensi (Copepoda : Caligidae) on three-spine stickleback Gasterosteus aculeatus in coastal British Columbia. J Parasitol. 2006, 92: 473-480. 10.1645/GE-685R1.1.
Pert CC, Mordue AJ, O’Shea B, Bricknell IR: The settlement and reproductive success of Lepeophtheirus salmonis (Kroyer 1837; Copepoda: Caligidae) on atypical hosts. Aquac Res. 2012, 43: 799-805. 10.1111/j.1365-2109.2011.02891.x.
Lyndon AR, Toovey JPG: Occurrence of gravid salmon lice (Lepeophtheirus salmonis (Kroyer)) on saithe (Pollachius virens (L.)) from salmon farm cages. Bull Eur Assn Fish Pathol. 2001, 21: 84-85.
Hamre LA, Glover KA, Nilsen F: Establishment and characterisation of salmon louse (Lepeophtheirus salmonis (Kroyer 1837)) laboratory strains. Parasitol Int. 2009, 58: 451-460. 10.1016/j.parint.2009.08.009.
Hamre LA, Nilsen F: Individual fish tank arrays in studies of Lepeophtheirus salmonis and lice loss variability. Dis Aquat Organ. 2011, 97: 47-56. 10.3354/dao02397.
Nolan DV, Martin SAM, Kelly Y, Glennon K, Palmer R, Smith T, McCormack GP, Powell R: Development of microsatellite PCR typing methodology for the sea louse, Lepeophtheirus salmonis (Kroyer). Aquac Res. 2000, 31: 815-822. 10.1046/j.1365-2109.2000.00514.x.
Nolan DV, Powell R: Geographic and temporal genetic structure in Lepeophtheirus salmonis from four salmon farms along the northwest and west coasts of Ireland: results from a microsatellite analysis. Hydrobiologia. 2009, 617: 55-63. 10.1007/s10750-008-9525-7.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S: MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011, 28: 2731-2739. 10.1093/molbev/msr121.
Tamura K, Nei M: Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993, 10: 512-526.
Lee CE: Global phylogeography of a cryptic copepod species complex and reproductive isolation between genetically proximate “populations”. Evolution. 2000, 54: 2014-2027.
Ganz HH, Burton RS: Genetic Differentiation and Reproductive Incompatibility among Baja-California Populations of the Copepod Tigriopus-Californicus. Mar Biol. 1995, 123: 821-827. 10.1007/BF00349126.
Grishanin AK, Rasch EM, Dodson SI, Wyngaard GA: Genetic architecture of the cryptic species complex of Acanthocyclops vernalis (Crustacea : Copepoda). II. Crossbreeding experiments, cytogenetics, and a model of chromosomal evolution. Evolution. 2006, 60: 247-256.
Pritchard VL, Dimond L, Harrison JS, Velazquez CCS, Zieba JT, Burton RS, Edmands S: Interpopulation hybridization results in widespread viability selection across the genome in Tigriopus californicus. BMC Genet. 2011, 12: 54-
Willett CS: Deleterious epistatic interactions between electron transport system protein-coding loci in the copepod Tigriopus californicus. Genetics. 2006, 173: 1465-1477. 10.1534/genetics.105.051011.
Glover KA, Hamre LA, Skaala O, Nilsen F: A comparison of sea louse (Lepeophtheirus salmonis) infection levels in farmed and wild Atlantic salmon (Salmo salar L.) stocks. Aquaculture. 2004, 232: 41-52. 10.1016/S0044-8486(03)00454-X.
Glover KA, Nilsen F, Skaala O: Individual variation in sea lice (Lepeophtheirus salmonis) infection on Atlantic salmon (Salmo salar). Aquaculture. 2004, 241: 701-709. 10.1016/j.aquaculture.2004.07.030.
Glover KA, Nilsen F, Skaala O, Taggart JB, Teale AJ: Differences in Susceptibility to Sea Lice Infection Between a Sea Run and a Freshwater Resident Population of Brown Trout. J Fish Biol. 2001, 59: 1512-1519. 10.1111/j.1095-8649.2001.tb00216.x.
Edmands S: Heterosis and outbreeding depression in interpopulation crosses spanning a wide range of divergence. Evolution. 1999, 53: 1757-1768. 10.2307/2640438.
Hebert PDN, Cywinska A, Ball SL, DeWaard JR: Biological identifications through DNA barcodes. Proc R Soc Lond Ser B-Biol Sci. 2003, 270: 313-321. 10.1098/rspb.2002.2218.
Hebert PDN, Ratnasingham S, deWaard JR: Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc Lond Ser B-Biol Sci. 2003, 270: S96-S99. 10.1098/rsbl.2003.0025.
Bucklin A, Guarnieri M, Hill RS, Bentley AM, Kaartvedt S: Taxonomic and systematic assessment of planktonic copepods using mitochondrial COI sequence variation and competitive, species-specific PCR. Hydrobiologia. 1999, 401: 239-254.
Lefebure T, Douady CJ, Gouy M, Gibert J: Relationship between morphological taxonomy and molecular divergence within Crustacea: Proposal of a molecular threshold to help species delimitation. Mol Phyl Evol. 2006, 40: 435-447. 10.1016/j.ympev.2006.03.014.
Parent GJ, Plourde S, Turgeon J: Natural hybridization between Calanus finmarchicus and C. glacialis (Copepoda) in the Arctic and Northwest Atlantic. Limnol Oceanol. 2012, 57: 1057-1066. 10.4319/lo.2012.57.4.1057.
Bucklin A, Lajeunesse TC: Molecular-genetic variation of calanus-pacificus (copepoda, calanoida) - preliminary evaluation of genetic-structure and subspecific differentiation based on mtdna sequences. Calif Coop Ocean Fish Invest Rep. 1994, 35: 45-51.
Nuwer M, Frost B, Armbrust EV: Population structure of the planktonic copepod Calanus pacificus in the North Pacific Ocean. Mar Biol. 2008, 156: 107-115.
Acknowledgements
We would like to acknowledge Stig Mæhle and Enrique Perez Garcia for technical assistance and in particular Dr. Egil Karlsbakk for constructive discussions. This study was jointly funded by the Norwegian Department for Fisheries and Coastal Affairs and the Norwegian research council project PrevenT.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors’ contributions
The study was conceived, designed and conducted by RSM, OT and KAG. The results were analyzed and the manuscript written by RSM and KAG. All authors read and approved the final manuscript.
Electronic supplementary material
12863_2013_1356_MOESM1_ESM.pdf
Additional file 1: Certificate from the Norwegian Food Safety Authority (NFSA) for import of Pacific L. salmonis from the Pacific Ocean.(PDF 759 KB)
12863_2013_1356_MOESM2_ESM.doc
Additional file 2: PCR amplification conditions for the 16 microsatellite markers used to validate maternal and paternal contribution to the F1 hybrid crosses.(DOC 124 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Skern-Mauritzen, R., Torrissen, O. & Glover, K.A. Pacific and Atlantic Lepeophtheirus salmonis (Krøyer, 1838) are allopatric subspecies: Lepeophtheirus salmonis salmonis and L. salmonis oncorhynchi subspecies novo. BMC Genet 15, 32 (2014). https://doi.org/10.1186/1471-2156-15-32
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2156-15-32