Abstract
Background
Soil-feeding termites are particularly interesting models for studying the effects of fragmentation, a natural or anthropic phenomenon described as promoting genetic differentiation. However, studying the link between fragmentation and genetics requires a method for identifying species unambiguously, especially when morphological diagnostic characters are lacking. In humivorous termites, which contribute to the fertility of tropical soils, molecular taxonomy and phylogenetic relationships are rarely studied, though mitochondrial and nuclear molecular markers are widely used in studies of pest termites. Here, we attempt to clarify the taxonomy of soil-feeding colonies collected throughout the naturally fragmented Lopé Reserve area (Gabon) and morphologically affiliated to Cubitermes sp. affinis subarquatus. The mitochondrial gene of cytochrome oxidase II (COII), the second nuclear rDNA internal transcribed spacer (ITS2) and five microsatellites were analyzed in 19 colonies.
Results
Bayesian Inference, Maximum Likelihood and Maximum Parsimony phylogenetic analyses, which were applied to the COII and ITS2 sequences, and Neighbor-Joining reconstructions, applied to the microsatellite data, reveal four major lineages in the Cubitermes sp. affinis subarquatus colonies. The concordant genealogical pattern of these unlinked markers strongly supports the existence of four cryptic species. Three are sympatric in the Reserve and are probably able to disperse within a mosaic of forests of variable ages and savannahs. One is limited to a very restricted gallery forest patch located in the North, outside the Reserve.
Conclusion
Our survey highlights the value of combined mitochondrial and nuclear markers for exploring unknown groups such as soil-feeding termites, and their relevance for resolving the taxonomy of organisms with ambiguous morphological diagnostic characters.
Similar content being viewed by others
Background
It has been suggested that ecosystem fragmentation has important consequences for animal populations by reducing and dividing the distribution areas and by limiting connections between the fragments [1, 2]. As a result, this natural or anthropological phenomenon is described as a process promoting the genetic differentiation of isolated populations and could be involved in speciation. Insects are interesting models for studying the effects of fragmentation owing to their abundance, limited dispersal, short generation time and sensitivity to disturbances. In tropical ecosystems, termites (Isoptera) may represent up to 95% of soil insect biomass; humivorous termites contribute to tropical soil fertilization [3–6]. Since soil-feeders are very sensitive to changes in their environment, they constitute interesting bio-indicators in landscape fragmentation studies, when the evolution of their specific richness is surveyed [7–9].
Studying the link between genetics and fragmentation requires evaluation of breeding structure, gene flow and genetic differentiation among populations. This implies unambiguous identification of species. For a long time, termite systematics was based on morphological and\or morphometrical character sets for individuals belonging to various castes (alates, soldiers or workers). During the past decade, an increasing number of taxonomic studies have shown that molecular methods constitute fast and reliable diagnostic systems, which complement morphological identification. Indeed, several studies based on mitochondrial genome sequences such as the cytochrome oxidase genes, the AT-rich region and the 16S rDNA have thrown a great deal of light on termite taxonomy and on phylogenetic and phylogeographic analyses of the Reticulitermes [10–15], Nasutitermes [16–18] and Heterotermes [19] genera. Because mitochondrial gene trees do not necessarily reflect species trees owing to their maternal inheritance, the addition of nuclear markers in molecular studies is useful for confirming the organismal phylogeny [20]. Among the nuclear sequences, the internal transcribed spacers (ITS) of rDNA are more polymorphic between than within species. It is generally assumed than concerted evolution homogenizes individual rDNA repeats and produces a mostly uniform sequence in all repeats in a given species. Although there is intra-individual variation in some taxa, the ITS2 region sequences are considered phylogenetically informative and able to distinguish closely-related species [21]. Such an evolutionary pattern has allowed sibling or cryptic species in the Reticulitermes genus to be discriminated [22–24]. Microsatellites are also very useful nuclear polymorphic markers and have contributed widely to the resolution of colony and population genetic structures in Isoptera [25–27]. Many surveys have concerned the xylophageous genus,Reticulitermes, in non-natural fragmented areas such as urban ecosystems [28, 29]. To date, however, microsatellite markers have not been used to investigate termite phylogeny or species discrimination, as has been done for other groups e.g. vertebrates [30], ticks [31], wasps [32] and ants [33].
In the present survey, we studied the humivorous afro-tropical Cubitermes genus (Termitidae, Termitinae), which shows remarkable ecological plasticity, colonizing savannahs or forests according to species. Among the soil-feeding Termitidae, the Cubitermes genus is one of the main builders; its nest can shelter up to 10 genera of true inquiline or optional builder termites [34]. In spite of its undeniable ecological interest, the taxonomy of this genus is poorly resolved and a complete revision appears necessary [35]. Indeed, at least 64 species of Cubitermes have been described on the basis of morphometrical characters, but it is strongly suspected that some of these are synonymic. It is important to note that extensive taxonomic work is necessary for the West African species since the sole currently available key concerns the East African species [36]. Molecular data are also lacking for the Cubitermes genus, as only one sequence from cytochrome oxidase I [37] and two from 12S mtDNA [GenBank: AF475037, AF475001] are registered.
The aim of this work is to clarify the taxonomic status of the Cubitermes colonies from the Lopé Reserve region (Gabon), in order to obtain a better understanding of tropical termite diversity in fragmented areas. The Lopé Reserve is typically characterized by a mosaic of forests and savannahs and constitutes an ideal setting for studies of natural fragmentation.
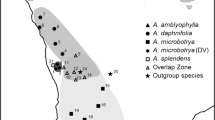
On the basis of morphological comparisons with type specimens in the collections of the British Museum (London) and the Royal Museum for Central Africa (Tervuren), and because they showed no diagnostic morphometrical variation or molecular divergence in mitochondrial 12S and 16S rDNA (Harry, unpublished data), the Cubitermes colonies were affiliated to a single species, Cubitermes sp. affinis subarquatus (Sjöstedt). Here we attempt to reconstruct a phylogeny based on three types of polymorphic and independent molecular markers. We sequenced a portion of the mitochondrial COII gene and the ITS2 region, and determined the genotypes at five microsatellite loci isolated from Cubitermes subarquatus [38], for 19 Cubitermes colonies from four different sites (Figure 1). Three of these sites were in the same geographical scale within the Lopé Reserve and corresponded to forest patches of different ages, including small savannah patches: Okoumé (75 years old), Rocher (800 years old) and Chameau (800 years old). A fourth site, Doda, was an isolated gallery-forest outside the Reserve within a savannah landscape.
Results
Mitochondrial sequence analyses
Mitochondrial sequence dataset consisted in 558 bp sequenced for one individual from each of the 19 colonies collected in the four sites, and for the outgroup taxa, Apilitermes longiceps and Crenetermes albotarsalis (Termitidae, Termitinae).
In total for the Cubitermes sequences, 42 nucleotidic sites were variable (7.5%) and the overall proportion of A+T reached 66.7%. Height haplotypes were scored (Table 1 and Figure 2) differing at 1–30 nucleotide sites (0.18–5.40% sequence divergence). Maximum Parsimony (MP), Bayesian Inference (BI) and Maximum Likelihood (ML) reconstructions clearly showed four distinct COII haplotype groups (Figure 3). We named the first clade Cubitermes spA, including sequences from the T7, T16, T42 and T14A colonies. The second clade, named Cubitermes spB, included T5, T26, T34, T37, T38 and T14B sequences. The third clade, Cubitermes spC, comprised T17, T24, T31, T33, T46, T45 and TX sequences and the fourth group, Cubitermes spD, only the two sequences from the colonies of the Doda site, TD1 and TD2.
Bayesian phylogenetic tree of mtDNA haplotypes. Bayesian tree showing relationships of COII sequences from Cubitermes sp. affinis subarquatus colonies. Apilitermes and Crenetermes sequences are used to root the tree. BI posterior probabilities followed by ML and MP bootstrap support values are indicated in bold at nodes. Putative species are indicated in bold on the right.
Sequence divergence was quite low within these groups. Indeed, in the Cubitermes spA group, only one haplotype was detected and sequence divergence was inferior to 1% within Cubitermes spB and Cubitermes spC groups (sequences differed by 0–0.18% and 0–0.36% respectively).
On the opposite, the divergence between the four mitochondrial lineages was high. The Cubitermes spA and Cubitermes spB sequences differed from each other by 3.94% (net sequence divergence), Cubitermes spA and Cubitermes spC by 4.22% and Cubitermes spB and Cubitermes spC by 4.99%. Finally, Cubitermes spD diverged by 1.07% from Cubitermes spA.
ITS2 sequence analyses
A total of 298 positions sequenced for one individual from each of the 19 Cubitermes colonies, for Apilitermes longiceps and Crenetermes albotarsalis were aligned for ITS2 sequences. Regarding Cubitermes sequences, we found 262 invariable sites, 25 alignment gaps and 11 polymorphic sites. MP, BI and ML reconstructions confirmed the four lineages found with the COII gene (Figure 4). However, the position of Cubitermes spD was quite different since it was not found univocally clustered with Cubitermes spA. In this ITS2 tree, high BI posterior probabilities, ML and MP bootstrap support values were found for grouping Cubitermes spA, C. spD and C. spC in the same clade (Figure 4, 99/68/100) whereas a lower resolution of this branch appeared in the tree of mtDNA haplotypes (Figure 3, 54/<50/<50).
Bayesian phylogenetic tree of ITS2 sequences. Bayesian tree showing relationships of ITS2 sequences from Cubitermes sp. affinis subarquatus colonies. Apilitermes and Crenetermes sequences are used to root the tree. BI posterior probabilities followed by ML and MP bootstrap support values are indicated in bold at nodes. Putative species are indicated in bold on the right.
Sequence data from ITS2 revealed no polymorphism within the putative four species. Since no heterozygote was detected, ITS2 sequences were assigned to four unique haplotypes (a for Cubitermes spA, b for Cubitermes spB, c for Cubitermes spC, and d for Cubitermes spD, Table 1).
Again, divergence between groups was high in comparison with within-group divergence. Haplotypes of Cubitermes spA differed from the Cubitermes spC haplotypes by 2.18% (net sequence divergence). The Cubitermes spA and Cubitermes spB haplotypes differed from each other by 7.61% and Cubitermes spB and Cubitermes spC by 8.36%. Finally, the haplotype of Cubitermes spD differed from the haplotype of Cubitermes spA by 1.43%.
Microsatellite analyses
A total of 447 individuals for the 19 nests were surveyed at five microsatellite loci. The genetic differentiation among colonies within each putative species (FCT = 0.275, 0.258, 0.322 for Cubitermes spA, C. spB and C. spC, respectively) was substantially low. Otherwise, very high genetic differentiation was detected between colonies of different Cubitermes putative species. Cubitermes spA and Cubitermes spD were the less differentiated (FSTAD= 0.12; CI = -0.09–0.38) followed by Cubitermes spA and Cubitermes spC groups (FSTAC= 0.25, CI = 0.11–0.39). Cubitermes spA and Cubitermes spB (FSTAB= 0.49, CI = 0.27–0.67) and Cubitermes spB and Cubitermes spC (FSTBC= 0.48, CI = 0.25–0.68) showed similar high patterns of differentiation.
The NJ trees based on the DAS distance, the minimum genetic distance of Nei and the chord distance (DC) showed a genetic structure with four main clusters (Figure 5).
Neighbor-Joining tree calculated from the microsatellite data. Distances between Cubitermes sp. affinis subarquatus colonies are calculated based on the chord distance of Cavalli-Sforza from five microsatellite loci. The tree is unrooted. Values at nodes represent bootstrap support values (only values >50% are shown). Putative species are indicated in bold.
Discussion
In tropical soil-feeding termites, it is impractical to apply a Biological Species Concept [39] because of the difficulties in realizing experimental crosses and observing natural hybridizations. Thus, our purpose here was to apply a Phylogenetic Species Recognition (PSR) based on the Genealogical Concordance Concept [40] to diagnose the Cubitermes species or to detect cryptic species. Such a concept has often been applied in bacteria, fungi and animals. In termites, research involving DNA-based taxonomy of structural and agricultural pest species is important [10]. Comparison of multiple molecular markers has allowed the taxonomic status of new species in Reticulitermes to be assessed [60, 61] and has provided evidence for species synonymy in this genus [12, 14, 23]. Many DNA-based species recognitions are corroborated by evidence of morphological/chemical differences or geographically defined groups. However, PSR can be a powerful tool for diagnosing otherwise undistinguishable species, because genetic changes occurring in recently-isolated species may be observed before morphological or behavioral changes arise [41]. As found recently in other social insects, genetic isolation is not always accompanied by evident morphological differentiation. In fire ants belonging to the Solenopsis genus, genetic analyses based on allozyme and mitochondrial markers demonstrated the occurrence of sympatric and indistinguishable cryptic species [42–44].
The genetic results obtained in our study from the combined mitochondrial, nuclear and microsatellite markers unequivocally show deep separation among four groups of genotypes in the Cubitermes sp. affinis subarquatus colonies from the Lopé Reserve region. The congruence of these three types of unlinked molecular markers strongly supports the existence of differentiated genetic pools in this limited area. The mitochondrial and nuclear sequences show very little variation within the four groups but there is considerable variation among them. Although the allele ranges overlap for all the loci, possibly because of size homoplasy, the microsatellite data confirm the occurrence of four differentiated groups. These unexpected levels of subdivision are unlikely to have arisen under gene flow, so these four newly-detected groups can be seen as good evidence for the existence of cryptic species previously unrecognized by morphological techniques.
The lack of detectable morphological differences among Cubitermes species is not surprising in view of the data in the taxonomic literature, which imply that the genus is highly complex (great number of species, synonymy, missing data, etc.) [35]. Furthermore, the boundaries between nominal Cubitermes species are often concealed by intra-specific morphometrical variability. In his revision of the East African Cubitermes species, Williams [36] mentioned that most of the specific characters vary greatly in size and can also vary markedly in shape and proportion within a particular species.
In addition, our genetic data inform the current debate about the use of comparative phylogenetic methods for studying present-day species distributions. These distributions do not necessarily reflect the geographical range of the ancestral species at the time of speciation, because geographical distributions are often labile owing to climate fluctuations, territory expansion or extinction of competitors. In particular, we have certainly not sampled the entire species range and enabled definite conclusions to be drawn about biogeographic differentiation and speciation. However, the repartition of the cryptic Cubitermes species is quite interesting in relation to the species distributions of poorly dispersive insects.
In our study, we can consider two landscape units: (1) the Lopé Reserve south of the Ogooué River, where Cubitermes spA, C. spB and C. spC live sympatrically in the closely apposed sites of Okoumé, Chameau and Rocher, the only exception being that Cubitermes spA is absent from the Chameau site; (2) the Doda zone to the north, where Cubitermes spD is restricted to the distant and isolated gallery-forest of Doda and represents the only Cubitermes species in the Doda zone across 10 km. The ecological differentiation of the two zones results from paleogeographic events. Indeed, since the last glaciations, the North Ogooué has progressively run dry, resulting in the regression of all forest types except riparian. In the South Ogooué, a wet environment has been maintained by the well-developed hydro-geographical network and this has allowed the forest to be preserved.
The contemporary sympatric distributions of three of the Cubitermes species (Cubitermes spA, C. spB and C. spC) in the Reserve zone could reflect their ability to disperse within a mosaic of forests of variable ages (e.g. differing in biotic and edaphic parameters) and savannah "buffer-zones". Indeed, their dispersion seems not to be affected by fragmentation on this small scale. Little is known about the dispersion modalities and the reproductive strategies of the Cubitermes species. It is very likely that swarming (i.e. alate dispersal flight) is the main mode of dispersal in this genus, since budding (i.e. local secondary reproduction initiated by the differentiation of neotenic reproductives, derived from the nymphs or workers of the colony) is not as common in Termitidae as in lower termites [45]. It has been suggested that although the active flight of winged termites is very limited (a few hundreds of meters), the meteorological conditions accompanying dispersal flights could strongly influence the distance covered by these sexual alates. Actually, a recent genetic study of Macrotermes michaelseni (Termitidae) suggested that some winged termites can travel considerable distances (50 km), most likely by passive drift [46]. Furthermore, the effectiveness of dispersal obviously depends upon the number of alates and the rate of predation [47].
Finally, the particular distribution pattern of Cubitermes spD raises the question of the link between the history of the forest fragmentation and the modalities of speciation. One can indeed wonder whether the history of successive modifications of habitats in this geographical zone has not contributed to the isolation of the termite species populating it. Areas such as isolated gallery forests, like Doda, may constitute refuges for fauna, where some species are led to disappear while others begin to differentiate under the influence of genetic drift.
Conclusion
The combination of mitochondrial and nuclear markers provides a reliable diagnostic method for separating Cubitermes species and offers a complement to the morphometrical diagnostic. Furthermore, these molecular markers could reveal useful information about their phylogenetical relationships. Similarly, such methods could be extended to termites for which species taxonomy is ambiguous.
Methods
Study site and species
The field collections were carried out in the Lopé Reserve region (Middle Ogooué, Gabon). The Lopé Reserve is constituted by a mosaic of forest and savannah, primarily formed during the last glaciations (-18000, -12000) and then maintained by human savannah burnings, resulting in a fragmented landscape. The sample area was composed of three forest sites in the Reserve, characterized by vegetation age and 2–5 km apart (Okoumé, Chameau and Rocher). The Okoumé site consisted of 75 years-old Milletia, Aucoumea and Marantacea stands. The Rocher and Chameau sites were older (800 years-old) and consisted in mature forest scattered by rock outcrops and classical pluvial forest stands, respectively. Finally, a fourth sampled site, Doda, was an isolated gallery-forest in a savannah landscape, dating up to 2500 years and situated outside the Reserve. In total, 19 colonies of Cubitermes sp. affinis subarquatus and two colonies of Apilitermes longiceps and Crenetermes albotarsalis were sampled (Table 1). Immediately following collection, individuals were placed in absolute ethanol and stored at 4°C in the laboratory until DNA extraction.
DNA extraction
Total genomic DNA from Cubitermes, Apilitermes and Crenetermes individuals was isolated using an extraction method with Wilson buffer (Tris hydrogen chloride 1 M, ethylenediaminetetraacetic acid 0.5 M, sodium chloride 4.5 M, sodium dodecylsulfate 20%, dithiothreitol, proteinase K) followed by a salting-out procedure.
Mitochondrial and ITS2 sequence analyses
DNA analyses were based on sequences from partial mitochondrial cytochrome oxidase subunit II gene (COII) and nuclear internal transcribed spacer 2 (ITS2). PCR was performed for one individual per colony (N = 19 Cubitermes + 1 Apilitermes + 1 Crenetermes) in a total volume of 40 μL (50 μL for ITS2), composed of 20 μL (25 μL) of Taq PCR Master Mix (Qiagen), 1.6 μL (4 μL) of each primer (10 pM), 15.2 μL (18.5 μL) of distilled water and 1.6 μL (2.5 μL) of template DNA. Primers for COII amplification were forward modified A-tLeu 5'-CAGATAAGTGCATTGGATTT-3' and reverse B-tLys 5'-GTTTAAGAGACCAGTACTTG-3' [48, 49], modified by Miura et al. [16]. ITS2 sequences were amplified using forward ITS2F 5'-TGTGAACTGCAGGACACAT-3' and reverse ITS2Rcub 5'-ATTCGGCGGGTAGTCTCG-3' primers modified in this study from Jenkins et al. [23]. The amplification conditions were adapted from Miura et al. [16]. The amplification products were purified using a DNA and Gel band Purification kit (GFX TM PCR kit, Amersham Biosciences USA). Sequence reactions were performed using BigDye Terminator Cycle Sequencing kit version 1.1 (Applied Biosystems), then purified with an ethanol-Na acetate method. Sequence data were obtained using an automatic DNA sequencer (Applied Biosystems, ABI PRISM 310) and analysed with Sequencing Data software (Applied Biosystems). All sequences were registered in GenBank database with accession numbers listed in Table 1.
COII and ITS2 sequences were aligned using CLUSTALW with the default settings [50] and sequences from Apilitermes longiceps and Crenetermes albotarsalis (Termitidae, Termitinae) were added to both datasets in order to root the trees. Phylogenetic analyses were performed using Maximum Parsimony (MP), Bayesian Inference (BI) and Maximum Likelihood (ML) methods.
MP trees were constructed using Phylip (Phylogeny Inference Package) Version 3.572 [51] with the SEQBOOT, DNAPARS and CONSENSE programs with 1000 repetitions of bootstrap.
For probabilistic methods, a nucleotide substitution model was selected for each sequence dataset using the Akaike Information Criterion (AIC) implemented in Modeltest v3.7 [52]. The best-fit substitution model selected was TrN+G (Nst = 6, Rates = gamma, Pinvar = 0) for COII sequences and TVM (Nst = 6, Rates = equal, Pinvar = 0) for ITS2 sequences. BI trees were constructed using MrBayes 3.1 [53]. We ran four Markov chains (one cold, three heated) for 1,000,000 generations, sampled every 100 generations (burnin = 2500, according to the convergence diagnostic). Consensus trees were generated including posterior probability of clades and branch lengths. ML trees were constructed using thePAUP 4.0b10 program [54] and the reliability of the inferred trees was tested by 100 bootstrap resamplings for COII and 40 for ITS2. Tree topologies were congruent across all methods and therefore, only BI trees are reported with posterior probabilities and bootstrap support values for ML and MP.
Microsatellite analyses
The genotypes for 18 to 48 sterile individuals from each colony except two (T14A, n = 6 and T14B, n = 12) i.e. a total of 447 individuals genotyped, were assayed at five microsatellite loci (P14, P19, P32, P34 and P41) by means of Polymerase Chain Reaction (PCR) amplification. Primer sequences and amplification conditions are given in Harry et al. [38]. PCR products were electrophoresed on an ABI Prism 310 DNA sequencer (Applied Biosystems) and microsatellite allele sizes were scored using the GENSCAN and GENOTYPER programs (Applied Biosystems).
We investigated whether genetic differentiation occurs among groups detected in the phylogenetic analyses. In order to do this, we estimated FST coefficients with individuals nested in colonies and colonies nested in major lineages using a three-level hierarchy in the GDA program [55], for each pair of putative cryptic species. 95% CIs were constructed by bootstrapping over loci with 1000 replications. Values for which 95% CIs did not overlap zero were considered as significantly different at the 0.05 level. We compared the allele differentiation among colonies of the same genetic group and among colonies of the same geographical site using the FCT coefficient [56] calculated with a two-level hierarchy in the GDA program.
Phylogenetic distances between colonies were estimated with the Populations 1.2.00 program [57] by using the genetic distance of shared alleles, the minimum genetic distance of Nei and the chord distance of Cavalli-Sforza. The resulting genetic distances and bootstrapping procedures (1000 replicates) were used to construct an unrooted consensus tree. Tree topologies were congruent across the three methods and therefore, only one reconstruction is presented here (reconstruction based on the chord distance of Cavalli-Sforza).
References
Keller I, Nentwig W, Largiader CR: Recent habitat fragmentation due to roads can lead to significant genetic differentiation in an abundant flightless ground beetle. Mol Ecol. 2004, 13: 2983-2994. 10.1111/j.1365-294X.2004.02310.x.
Krauss J, Schmitt T, Seitz A, Steffan-Dewenter I, Tscharntke T: Effects of habitat fragmentation on the genetic structure of the monophagous butterfly Polyommatus coridon along its northern range margin. Mol Ecol. 2004, 13: 311-320. 10.1046/j.1365-294X.2003.02072.x.
Garnier-Sillam E, Harry M: Distribution of humic compounds in mounds of some soil-feeding termite species of tropical rainforests: its influence on soil structure stability. Insectes Soc. 1995, 42: 167-185. 10.1007/BF01242453.
Fall S, Brauman A, Chotte JL: Comparative distribution of organic matter in particle and aggregate size fractions in the mounds of termites with different feeding habits in Senegal: Cubitermes niokoloensis and Macrotermes bellicosus. Agric, Ecosyst Environ, Appl Soil Ecol. 2001, 17: 131-140.
Lobry de Bruyn LA, Conacher AJ: Soil modification by termites in the central wheat-belt of Western-Australia. Aust J Soil Res. 1995, 33: 179-193. 10.1071/SR9950179.
Wood TG: Termites and the soil environment. Biol Fertil Soils. 1988, 6: 228-236. 10.1007/BF00260819.
Eggleton P, Bignell DE, Hauser S, Dibog L, Norgrove L, Madong B: Termite diversity across an anthropogenic disturbance gradient in the humid forest zone of West Africa. Agric, Ecosyst Environ, Appl Soil Ecol. 2002, 90: 189-202. 10.1016/S0167-8809(01)00206-7.
Eggleton P, Bignell DE, Sands WA, Mawdsley NA, Lawton JH, Wood TG, Bignell NC: The diversity, abundance and biomass of termites under differing levels of disturbance in the Mbalmayo Forest Reserve, southern Cameroon. Philos Trans R Soc Lond, B, Biol Sci. 1996, 351: 51-68.
Fonseca de Souza OF, Brown VK: Effects of habitat fragmentation on Amazonian termite communities. J Trop Ecol. 1994, 10: 197-206.
Austin JW, Szalanski AL, Gold RE, Foster BT: DNA-based identification of the Eastern subterranean termite Reticulitermes flavipes (Isoptera: Rhinotermitidae). J Econ Entomol. 2004, 97: 95-101.
Austin JW, Szalanski AL, Gold RE, Foster BT: Genetic variation and geographical distribution of the subterranean termite genus Reticulitermes in Texas. Southwest Entomol. 2004, 29: 1-12.
Austin JW, Szalanski AL, Scheffrahn RH, Messenger MT, Dronnet S, Bagnères AG: Genetic evidence for the synonymy of two Reticulitermes species: Reticulitermes flavipes and Reticulitermes santonensis. Ann Entomol Soc Am. 2005, 98: 395-401. 10.1603/0013-8746(2005)098[0395:GEFTSO]2.0.CO;2.
Luchetti A, Trenta M, Mantovani B, Marini M: Taxonomy and phylogeny of north mediterranean Reticulitermes termites (Isoptera, Rhinotermitidae): a new insight. Insectes Soc. 2004, 51: 117-122. 10.1007/s00040-003-0715-z.
Marini M, Mantovani BUR: Molecular relationships among European samples of Reticulitermes (Isoptera, Rhinotermitidae). Mol Phylogenet Evol. 2002, 22: 454-459. 10.1006/mpev.2001.1068.
Clément JL, Bagnères AG, Uva P, Wilfert L, Quintana A, Reinhard J, Dronnet S: Biosystematics of Reticulitermes termites in Europe: morphological, chemical and molecular data. Insectes Sociaux. 2001, 48: 202-215.
Miura T, Roisin Y, Matsumoto T: Molecular phylogeny and biogeography of the nasute termite genus Nasutitermes (Isoptera: Termitidae) in the pacific tropics. Mol Phylogenet Evol. 2000, 17: 1-10. 10.1006/mpev.2000.0790.
Scheffrahn RH, Jkrecek J, Szalanski AL, Austin JW: Synonymy of neotropical arboreal termites Nasutitermes corniger and N. costalis (Isoptera: Termitidae: Nasutitermitinae), with evidence from morphology, genetics, and biogeography. Ann Entomol Soc Am. 2005, 98: 273-281. 10.1603/0013-8746(2005)098[0273:SONATN]2.0.CO;2.
Scheffrahn RH, Krecek J, Szalanski AL, Austin JW, Roisin Y: Synonymy of two arboreal termites (Isoptera:Termitidae: nasutitermitinae): Nasutitermes corniger from the Neotropics and N. polygynus from New Guinea. Fla Entomol. 2005, 88: 28-33. 10.1653/0015-4040(2005)088[0028:SOTATI]2.0.CO;2.
Szalanski AL, Scheffrahn RH, Austin JW, Krecek J, Su NY: Molecular phylogeny and biogeography of Heterotermes (Isoptera: Rhinotermitidae) in the West Indies. Ann Entomol Soc Am. 2004, 97: 556–566-10.1603/0013-8746(2004)097[0556:MPABOH]2.0.CO;2.
Arevalo E, Zhu Y, Carpenter J, Strassmann J: The phylogeny of the social wasp subfamily Polistinae: evidence from microsatellite flanking sequences, mitochondrial COI sequence, and morphological characters. BMC Evol Biol. 2004, 4: 8-10.1186/1471-2148-4-8.
Alvarez JM, Hoy MA: Evaluation of the ribosomal ITS2 DNA sequences in separating closely related populations of the parasitoid Ageniaspis (Hymenoptera: Encyrtidae). Ann Entomol Soc Am. 2002, 95: 250-256. 10.1603/0013-8746(2002)095[0250:EOTRID]2.0.CO;2.
Uva P, Clement JL, Austin JW, Aubert J, Zaffagnini V, Quintana A, Bagneres AG: Origin of a new Reticulitermes termite (Isoptera, Rhinotermitidae) inferred from mitochondrial and nuclear DNA data. Mol Phylogenet Evol. 2004, 30: 344-353. 10.1016/S1055-7903(03)00202-1.
Jenkins TM, Dean RE, Verkerk R, Forschler BT: Phylogenetic analyses of two mitochondrial genes and one nuclear intron region illuminate European subterranean termite (Isoptera: Rhinotermitidae) gene flow, taxonomy, and introduction dynamics. Mol Phylogenet Evol. 2001, 20: 286-293. 10.1006/mpev.2001.0966.
Kutnik M, Uva P, Brinkworth L, Bagneres AG: Phylogeography of two European Reticulitermes (Isoptera) species: the Iberian refugium. Mol Ecol. 2004, 13: 3099-3113. 10.1111/j.1365-294X.2004.02294.x.
Hacker M, Kaib M, Bagine RKN, Epplen JT, Brandl R: Unrelated queens coexist in colonies of the termite Macrotermes michaelseni. Mol Ecol. 2005, 14: 1527-1532. 10.1111/j.1365-294X.2005.02507.x.
Vargo EL: Hierarchical analysis of colony and population genetic structure of the eastern subterranean termite, Reticulitermes flavipes, using two classes of molecular markers. Evolution. 2003, 57: 2805-2818. 10.1554/03-336.
Vargo EL, Husseneder C, Grace JK: Colony and population genetic structure of the Formosan subterranean termite, Coptotermes formosanus, in Japan. Mol Ecol. 2003, 12: 2599-2608. 10.1046/j.1365-294X.2003.01938.x.
Dronnet S, Chapuisat M, Vargo EL, Lohou C, Bagnères AG: Genetic analysis of the breeding system of an invasive subterranean termite, Reticulitermes santonensis, in urban and natural habitats. Mol Ecol. 2005, 14: 1311-1320. 10.1111/j.1365-294X.2005.02508.x.
Vargo EL: Genetic structure of Reticulitermes flavipes and R. virginicus (Isoptera: Rhinotermitidae) colonies in an urban habitat and tracking of colonies following treatment with Hexaflumuron bait. Environ Entomol. 2003, 32: 1271-1282.
Richard M, Thorpe RS: Can microsatellites be used to infer phylogenies? Evidence from population affinities of the Western Canary Island lizard (Gallotia galloti). Mol Phylogenet Evol. 2001, 20: 351-360. 10.1006/mpev.2001.0981.
Mccoy KD, Boulinier T, Tirard C, Michalakis Y: Host specificity of a generalist parasite: genetic evidence of sympatric host races in the seabird tick Ixodes uriae. J Evol Biol. 2001, 14: 395-405. 10.1046/j.1420-9101.2001.00290.x.
Molbo D, Krieger MJB, Herre EA, Keller L: Species-diagnostic microsatellite loci for the fig wasp genus Pegoscapus. Mol Ecol Notes. 2002, 2: 440-442. 10.1046/j.1471-8286.2002.00268.x.
Macaranas JM, Colgan DJ, Major RE, Cassis G, Gray MR: Species discrimination and population differentiation in ants using microsatellites. Biochem Syst Ecol. 2001, 29: 125-136. 10.1016/S0305-1978(00)00038-7.
Dejean A, Ruelle JE: Importance of Cubitermes termitaries as shelter for alien incipient termite societies. Insectes Soc. 1995, 42: 129-136. 10.1007/BF01242449.
Ruelle JE: The genus Cubitermes Wasmann (Isoptera: Termitidae, Termitinae): a review of its taxonomy, nomenclature and distribution in Africa. J Afr Zool. 1992, 106: 499-502.
Williams RMC: The East African termites of the genus Cubitermes (Isoptera: Termitidae). Trans Roy Entomol Soc London. 1966, 118: 73-118.
Aanen DK, Eggleton P, Rouland-Lefèvre C, Guldberg-Froslev T, Rosendahl S: The evolution of fungus-growing termites and their mutualistic fungal symbionts. Proc Natl Acad Sci USA. 2002, 99: 14887-14892. 10.1073/pnas.222313099.
Harry M, Roose CL, Garnier-Sillam E, Solignac M: Microsatellite markers in soil-feeding termites (Cubitermes subarquatus, Isoptera, Termitidae, Termitinae). Mol Ecol. 2001, 1: 226-228.
Mayr E: Animal species and evolution. 1963, Cambridge, Belknap Press of Harvard University Press
Avise JC, Ball RM: Principles of genealogical concordance in species concepts and biological taxonomy. Oxford Surveys in Evolutionary Biology. Edited by: Futuyma D and Antonovics J. 1990, , Oxford University Press, 45–67-
Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC: Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol. 2000, 31: 21-32. 10.1006/fgbi.2000.1228.
Ross KG, Trager JC: Systematics and population genetics of fire ants (Solenopsis saevissima complex) from Argentina. Evolution. 1990, 44: 2113-2134. 10.2307/2409620.
Ross KG, Shoemaker DD: Species delimitation in native South American fire ants. Mol Ecol. 2005, 14: 3419-3438. 10.1111/j.1365-294X.2005.02661.x.
Shoemaker DDW, Ahrens ME, Ross KG: Molecular phylogeny of fire ants of the Solenopsis saevissima species-group based on mtDNA sequences. Mol Phylogenet Evol. 2006, 38: 200-215. 10.1016/j.ympev.2005.07.014.
Myles TG: Review of secondary reproduction in termites (Insecta: Isoptera) with comments on its role in termite ecology and social evolution. Sociobiology. 1999, 33: 1-91.
Brandl R, Hacker M, Epplen JT, Kaib M: High gene flow between populations of Macrotermes michaelseni (Isoptera, Termitidae). Insectes Soc. 2005, V52: 344-349. 10.1007/s00040-005-0820-2.
Nutting WL: Flight and colony foundation. Biology of Termites. Edited by: Krishna K and Weesner FM. 1969, New York, Academic Press, 1: 233-282.
Liu H, Beckenbach AT: Evolution of the mitochondrial cytochrome oxidase II gene among 10 orders of insects. Mol Phylogenet Evol. 1992, 1: 41-52. 10.1016/1055-7903(92)90034-E.
Simon C, Frati F, Beckenback A, Crespi B, Liu H, Flook P: Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved PCR primers. Ann Entomol Soc Am. 1994, 87: 651-701.
Thompson JD, Higgins DG, Gibson TJ: CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22: 4673-4680.
Felsenstein J: PHYLIP (phylogeny inference package) version 3.5c. 1993, Seattle, Department of Genetics, University of Washington
Posada D, Crandall KA: Modeltest: testing the model of DNA substitution. Bioinformatics. 1998, 14: 817-818. 10.1093/bioinformatics/14.9.817.
Huelsenbeck JP, Ronquist F: MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001, 17: 754-755. 10.1093/bioinformatics/17.8.754.
Swofford DL: PAUP: Phylogenetic analysis using parsimony. Version 4.0 b10. 1997, Washington, DC, Smithsonian Institution, Swofford DL: PAUP* – Phylogenetic Analyses Using Parsinomyand other methods, version 4.0. In Book PAUP* – PhylogeneticAnalyses Using Parsinomy and other methods, version 4.0, 4.0 edCity: Sinauer; 2003.
Lewis PO, Zaykin D: Genetic Data Analysis: Computer program for the analysis of allelic data. Version 1.0 (d16c). 2001, [http://hydrodictyon.eeb.uconn.edu/people/plewis/software.php]
Thorne BL: Evolution of eusociality in termites. Annu Rev Ecol Syst. 1997, 28: 27-54. 10.1146/annurev.ecolsys.28.1.27.
Langella O: Populations, a free population genetic software. 1999, [http://bioinformatics.org/project/?group_id=84]
Christy P, Jaffré R, Ntougou O, Wilks C: La forêt et la filière bois au Gabon. 2003, Multipress-Gabon, 389-
White L, Abernethy K: Guide de la végétation de la réserve de la Lopé Gabon. 1996, Libreville (Gabon), Multipress Gabon, 224-
Uva P, Clement J-L, Austin JW, Aubert J, Zaffagnini V, Quintana A, Bagneres A-G: Origin of a new Reticulitermes termite (Isoptera, Rhinotermitidae) inferred from mitochondrial and nuclear DNA data. Mol Phylogenet Evol. 2004, 30: 344-353. 10.1016/S1055-7903(03)00202-1.
Clement J-L: Enzymatic polymorphism in the European populations of various Reticulitermes species (Isoptera). Biosystematics of Social Insects. 1981, 49-61. Edited by Howse PE, Clément J-L. London: Academic Press
Acknowledgements
We are grateful to the authorities of Gabon represented by Mr E. Mamfoumbi of the 'Ministère des Eaux et Forêts' for permission to work in the Lopé Reserve. We thank K. Abernethy and L. White for field facilities and scientific collaboration. We also would like to thank J. Harris and M. E. Cruz de Carvalho for editing the English text and anonymous reviewers for helpful comments on the manuscript. Partial financial support of the ABI Prism automated sequencer came from the University of Paris XII – Val de Marne via BQR grant.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
VR and CD carried out the molecular genetic studies, participated at the data analysis and the manuscript draft. AL intensively helped VR and CD in DNA sequencing. MH initiated the study, realized the sampling, contributed to the analysis of the results and to the writing of the paper. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Roy, V., Demanche, C., Livet, A. et al. Genetic differentiation in the soil-feeding termite Cubitermes sp. affinis subarquatus: occurrence of cryptic species revealed by nuclear and mitochondrial markers. BMC Evol Biol 6, 102 (2006). https://doi.org/10.1186/1471-2148-6-102
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2148-6-102