Abstract
Background
Desert hedgehog (DHH) belongs to the hedgehog gene family that act as secreted intercellular signal transducers. DHH is an essential morphogen for normal testicular development and function in both mice and humans but is not present in the avian lineage. Like other hedgehog proteins, DHH signals through the patched (PTCH) receptors 1 and 2. Here we examine the expression and protein distribution of DHH, PTCH1 and PTCH2 in the developing testes of a marsupial mammal (the tammar wallaby) to determine whether DHH signalling is a conserved factor in gonadal development in all therian mammals.
Results
DHH, PTCH1 and PTCH2 were present in the marsupial genome and highly conserved with their eutherian orthologues. Phylogenetic analyses indicate that DHH has recently evolved and is a mammal-specific hedgehog orthologue. The marsupial PTCH2 receptor had an additional exon (exon 21a) not annotated in eutherian PTCH2 proteins. Interestingly we found evidence of this exon in humans and show that its translation would result in a truncated protein with functions similar to PTCH1. We also show that DHH expression was not restricted to the testes during gonadal development (as in mice), but was also expressed in the developing ovary. Expression of DHH, PTCH1 and PTCH2 in the adult tammar testis and ovary was consistent with findings in the adult mouse.
Conclusions
These data suggest that there is a highly conserved role for DHH signalling in the differentiation and function of the mammalian testis and that DHH may be necessary for marsupial ovarian development. The receptors PTCH1 and PTCH2 are highly conserved mediators of hedgehog signalling in both the developing and adult marsupial gonads. Together these findings indicate DHH is an essential therian mammal-specific morphogen in gonadal development and gametogenesis.
Similar content being viewed by others
Background
Desert hedgehog (DHH) is a member of the hedgehog gene family which act as secreted intercellular signal transducers [1]. Hedgehog was first identified as a segment polarity gene in Drosophila and has since been identified as a key regulator of pattern formation in embryonic and adult development in many vertebrate and invertebrate species. In addition, the hedgehog gene has undergone duplications in both invertebrates and vertebrates to produce a number of orthologues [2–6]. Mammals have three hedgehog orthologues, Sonic (Shh), Indian (Ihh), and desert (Dhh) hedgehog [3, 4, 7–9]. All three share a striking homology with the Drosophila orthologue [10]. Each mammalian orthologue has a unique, largely non-overlapping expression pattern, except in the gut where Ihh and Shh are co-expressed, and in the adult ovaries where Ihh and Dhh are co-expressed [11, 12]. Shh has an essential role in early fetal development, and is required for correct formation of the limbs, phallus, somites and neural tube [8–10]. Ihh has a more restricted developmental role, and is essential for chondrocyte development [13]. Dhh is essential for the maintenance of the male germ line and spermatogenesis [14]. Dhh is also expressed in Schwann cells, and appears to play a role in nerve sheath formation [15, 16].
Hedgehog actions are mediated at the cell surface by a multi-component receptor complex comprising the patched (PTCH) receptors and smoothened (SMO) protein [17]. Both proteins have orthologues in Drosophila that are also involved in hedgehog signal transduction and pattern formation. Initially, all three hedgehog protein functions were thought to be mediated through the PTCH1 receptor in mammals. However, a second orthologue was identified, PTCH2, which shares significant sequence homology with PTCH1. PTCH1 and PTCH2 both bind all hedgehog family members with similar affinities, and each forms a complex with SMO. However, the expression patterns of PTCH1 and PTCH2 do not entirely overlap, suggesting some degree of functional specialization [18]. While PTCH1 is widely expressed throughout the mouse embryo, PTCH2 is most predominant in the skin and in testis [18].
In the mouse, Dhh is expressed in the presumptive testis from E11.5 through to adult stages. Dhh was initially thought to be specifically expressed in the testis in the pre-Sertoli cells and so it was suggested it may be directly modulated by Sry [1, 10, 19] or its downstream partner Sox9 [20–22]. Dhh-null male mice are sex reversed [21, 23], their gonads are small and devoid of sperm and the mice develop as phenotypic females due to a lack of male steroid hormone production [20, 21]. Leydig cell numbers are dramatically decreased in the Dhh null gonad, but are not entirely absent [24]. Dhh appears to be an important regulator of Leydig cell development since its over-expression in the somatic cells can induce Leydig cell development [25]. In contrast to the Dhh-null males, female mice lacking the Dhh gene develop normally and are fertile [1]. While Dhh does not appear to be important for ovarian development, Dhh mRNA has been detected in the granulosa cells of preantral and antral follicles suggesting it can be activated in an SRY-independent manner [1, 12, 24–26]. In humans, mutations in DHH cause a range of phenotypes in 46, XY male patients from mild [27] to complete gonadal dysgenesis, including bilateral streak gonads, normally developed Müllerian ducts, and female external genitalia [28].
The majority of genes involved in testicular differentiation are highly conserved among the vertebrates [29]. However, no DHH orthologue has been identified in birds, although a paralogue (annotated as DHH) is present in the zebrafish (Danio), anole lizard (Anolis) and African clawed frog (Xenopus) genomes http://www.ensembl.org. Here we describe high conservation of DHH, PTCH1 and 2 in the genome of a marsupial, the tammar wallaby (Macropus eugenii). Marsupials, have been evolving independently of eutherian mammals for 130- 148 million years [30]. To determine when DHH evolved its role in mammalian testicular development, we examined the expression of DHH and its receptors PTCH1 and 2 during marsupial gonad development.
Methods
Animals
Tammar wallabies of Kangaroo Island, South Australia origin were maintained in our breeding colony in open grassy enclosures. Husbandry, handling and experiments were in accordance with the National Health and Medical Research Council of Australia/Commonwealth Scientific and Industrial Research Organization/Australian Research Council (1990) guidelines and approved by the University of Melbourne Animal Experimentation Ethics Committees.
Pregnancies were initiated by removal of the pouch young (RPY) from animals carrying a blastocyst in embryonic diapause [31, 32]. The day of RPY is designated Day 0 of pregnancy and births occur 26-27 days later [32]. Fetuses were removed from the uterus, dissected and tissues snap frozen in liquid nitrogen for mRNA expression analyses or fixed in 4% paraformaldehyde overnight at 4°C for protein localization. The sex of the fetus or pouch young was determined by the presence of scrotal bulges in males or mammary primordia in females [33] or by PCR for SRY [34]. The developing gonadal ridge first becomes apparent around day 22 of gestation in the fetal tammar wallaby [35]. We examined gonads from day 23 of gestation through to adulthood in both males and females.
Cloning of a tammar wallaby Dhh, PTCH1 and PTCH2
A full length DHH cDNA clone was isolated from a lambda-Zap II cDNA library (constructed from total RNA combined from male and female day 5 postpartum pouch young). The cDNA library was constructed by Clontech Laboratories. Libraries were plated to a density of 120, 000 plaque-forming units per 22 × 22 cm Nunc plate. The library was screened with a full-length mouse Dhh cDNA probe at 60°C in Church's buffer [36].
PTCH1 and 2 sequences were partially isolated from the tammar wallaby genome trace-archives http://www.ensembl.org. PCR and RACE was used to fully characterise the open reading frame (primers listed in Additional file 1) of each receptor. PCR fragments were cloned into p-GEM-T-Easy plasmid (Promega) and sequenced to verify. RACE was performed according to manufacturers instructions (Roche).
Phylogenetic analyses
Complete cDNA sequences for annotated, full-length hedgehog and patched family members were obtained from the Ensembl database (Ensembl identification numbers listed in Additional file 2) and aligned using MUSCLE http://www.ebi.ac.uk. Trees with fewer than 20 nodes were constructed by aligning sequences using MUSCLE and assembling tree with Mr.Bayes 3.1.2 at 100, 000 generations, sampled every 100 with burnin set to the first 250. Larger trees were aligned with TCoffee http://www.ebi.ac.uk and assembled with Mr.Bayes 3.1.2 at 1, 000, 000 generations, sampled every 1000 with burnin set to the first 250. All trees were viewed and ordered in FigTree 1.3.1.
RNA extraction
Total RNA was extracted from frozen tissues using the GenElute Total RNA mini kit (Sigma). RNA was DNase treated using the DNAfree reagent (Ambion). DNA free RNA was reverse transcribed using a dT primer and the superscript III kit according to manufacturers instructions (Invitrogen). All RT-PCR derived products were sequence verified as described above.
Immunohistochemistry
Tissues were fixed in 4% PFA overnight and embedded in paraffin wax according to standard methods. Deparaffinised tissue sections (8 μm) were treated with 3% hydrogen peroxide in methanol for 5 min to quench endogenous peroxidase activity. Antigen retrieval was achieved by placing slides in boiling 0.05 M Tris HCL pH 9.0 for 20 min. Primary antibodies were applied to sections as follows; DHH (anti-mouse DHH, goat polyclonal IgG, R&D Systems, cat.#AF196), 1:400, PTCH1 (anti-human PTCH1, rabbit polyclonal IgG, Abcam, cat.#AB39266) 1:400 and PTCH2 (anti-human PTCH2, rabbit polyclonal IgG, Lifespan BioSciences, cat#LS-B301) 1:400. The DHH antibody was verified by Western Blot to detect a single protein of 43 kDa (the predicted size of the tammar wallaby DHH protein; result not shown Additional file 3). Furthermore, DHH is the primary hedgehog ligand expressed in the marsupial gonad [37]. While the PTCH1 and 2 antibodies were not validated by western blot in the tammar wallaby, the PTCH1 and PTCH2 antibodies were raised against epitopes that share 99 and 100% homology respectively with the analogous tammar target proteins, but that show no cross reactivity. In addition, no other homologous sequences exist within the tammar genome with which the antibody could non-specifically cross react [38]. Primary antibodies were incubated at 4°C overnight. Depending on the primary antibody, the secondary antibody was either an anti-goat IgG raised in rabbit (Millipore) or an anti-rabbit IgG raised in goat (Santa Cruz Biotechnology), both conjugated to biotin and used at a 1:250 dilution. Signal was amplified using the ABC kit (Vector Labs) and visualised using 3-amino-9-ethylcarbazole (AEC) (Vector Labs) and sections were counterstained with haematoxylin. Immunohistochemistry was performed on at least 3 independent samples at each stage. Negative controls were performed as above with the primary antibody omitted.
Fluorescence In SituHybridisation Mapping
Chromosomes were prepared from peripheral blood according to standard methods [39]. Chromosome in situ suppression (CISS) hybridisation of the genomic lambda clones was performed, with minor modifications [40]. The largest DHH cDNA clone was labelled with digoxigenin using nick translation. The labelled probe was incubated with 50 ng of tammar wallaby Cot1 DNA before hybridisation to tammar-fixed metaphase chromosome spreads for 48 h at 37°C. Slides were washed in 0.2 × SSC at 60°C and hybridisation detected with anti-DIG-mouse antibody (Serva), followed by tetramethyl rhodamine iso-thiocyanate (TRITC) conjugated goat-anti-mouse antibody (Serva). Following hybridisation, the slides were counterstained with DAPI (4, 6-diamidino-2-phenylindole) to visualise the chromosomes.
Expression analyses
RT-PCR was carried out according to standard methods using oligo dT primed cDNA (superscript III, Invitrogen) and 30 PCR cycles. No-template negative controls were included in each round, and in each case showed no amplification (data not shown). A single sample was used for each amplification.
Quantitative PCR was similarly performed on oligo dT primed, reverse transcribed mRNA was prepared as outlined above. Quantitative PCR was carried out using the IQ Sybr green master mix (BioRad) in 20 ul reactions using primers for tammar DHH, PTCH1 and PTCH2 (Table 1) and normalised to β-actin as previously described [41]. Each PCR was performed on a single sample in triplicate (except for the adult ovary which was verified on three independent samples) and only those samples with a standard deviation of less than 1 cycle considered for analyses. Relative changes in gene expression were analysed using the methods described by Pfaffl [39, 42]. The variance in cycle threshold (ΔCt) was normalised across samples (with beta actin levels set to 100%) and inversely plotted as a percentage of the earliest ΔCt.
Results
Isolation of the tammar wallaby DHHcDNA
Eight independent cDNA clones of DHH were isolated from the tammar wallaby pouch-young cDNA library. The two cDNA clones containing the longest 5' and 3' UTRs and overlapping with each other in the coding region were sequenced. The combined length of the two DHH clones was 2.38 kb with a predicted open reading frame of 1, 197 bp. This encoded a protein of 399 amino acids with a predicted molecular weight of 44 kD. There were several common features between the predicted tammar DHH protein and that of mouse [43]. A short stretch of N-terminal residues (25 for the tammar and 22 for the mouse) were highly hydrophobic and presumed to function as a signalling peptide. There was a conserved 6 amino acid stretch, CGPGRG, after the signal peptide used to generate the secreted form of DHH. Drosophila hedgehog protein and vertebrate Shh, and Dhh proteins are processed into two smaller secreted peptides by an auto-proteolytic process, both in vitro and in vivo [44]. The catalytic site, GCF, was conserved in the tammar DHH protein, suggesting a similar auto-proteolytic mechanism may also occur to produce a 19.9 kD N-terminal peptide (amino acids 1-200) and a 21.6 kD C-terminal peptide (amino acids 201-403). Furthermore, the tammar and mouse DHH proteins share 96% identity in the N-terminal peptide and 94.5% in the C-terminal peptide, implying that both regions have been highly conserved during evolution (Additional file 4).
PTCH1 and 2 were partially isolated from the tammar wallaby genome http://www.ensembl.org. PCR and RACE was used to fill in missing portions to complete the open reading frame for each gene. PTCH1 was seven amino acids longer than its human orthologue and shares 83% sequence homology and 96% amino acid similarity. Conservation was particularly high (85%-100%) in the transmembrane domains and there was complete conservation of the putative glycosylation sites (Additional file 5).
PTCH2 was more divergent, but still shared 79% sequence identity and 89% amino acid similarity with eutherian orthologues. The tammar PTCH2 predicted protein was 11 amino acids shorter at the N-terminus compared to eutherian PTCH2. In addition we identified a 70 amino acid contiguous insertion immediately downstream of the 12th (last) transmembrane domain located at the C-terminus, which was present in cDNA isoforms isolated from both the day 3 and day 14 post partum (pp) testis (Figure 1). A relaxed Blast search of the non-redundant protein and nucleotide database with this 70 amino acid (210 base pair) sequence failed to return any homology in any described species, except for hits to human and opossum PTCH2 in the same location, downstream of the terminal transmembrane domain. The insertion shows 70% nucleotide identity and 79% amino acid similarity with human. Interestingly, one of the base changes in the human sequence (corresponding to amino acid 35) produced a premature stop codon (Figure 1a). As a result, there were no human protein hits that included this region, only nucleotide homology, and no known ESTs mapping to the nucleotide stretch. The sequence maps to reads in the unannotated genome archives of the opossum with 81% nucleotide identity and 77% amino acid similarity. The sequence shows low homology (< 30% contiguous nucleotide similarity) to mouse and cow gDNA in the same region. Additional exon aside, the C-terminus was the most variable region both within and between eutherians and marsupials (Additional file 6a). As with PTCH1, the transmembrane domains and glycosylation sites were all extremely conserved with 90-100% sequence identity within these regions. Additionally, multiple truncated splice variants of PTCH2 showing developmentally-regulated expression were identified during the subcloning process (Additional file 6b).
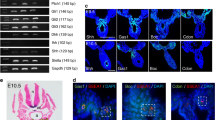
The structure and evolution of PTCH2 in therian mammals. a. Amino Acid alignment of the additional PTCH2 tammar wallaby (Me) exon 21a with sequence derived from intron 21 in humans (Hs). The red arrowhead indicates the location of the premature stop codon (-) at amino acid 35 in humans. Black shading indicates identical amino acids, grey shading indicates like amino acids and no shading indicates divergent amino acids. Numbers indicate amino acid position. b. Schematic of the PTCH2 protein derived from the native human isoform (left) the Δ-22 PTCH2 isoform (middle) and tammar PTCH2 with the additional exon 21a (right). c. Phylogenetic tree showing the evolution of exon 21a in mammals. Exon 21a was present in the common therian ancestor (blue arrowhead). The exon was retained without a nonsense codon in the marsupial lineage (blue arrowhead), was mutated in humans to contain a premature stop resulting in the Δ-22 isoform (red arrowhead) and was lost in the rodent lineage (green arrowhead).
Phylogenetic relationships of DHH, PTCH1 and PTCH2 to eutherian and non-mammalian vertebrates
A phylogenetic analysis of DHH sequences grouped them into four main clusters, one for primates, another elephant, mouse, and hyrax, a third for pig, dog, and dolphin; tammar was an outlier (Additional file 7a). These groupings are largely consistent with the accepted mammalian phylogenetic tree and all eutherian clusters show equal divergence.
DHH orthologues are absent from the bird genome, but are annotated in other non-mammalian vertebrates including the anole lizard, African clawed frog, zebrafish, medaka, stickleback and puffer fish. To determine the evolutionary relationship of these orthologues to mammalian DHH we constructed a phylogenetic tree (as described above) using complete sequences for SHH, IHH and DHH for all vertebrate species, as well as for the zebrafish specific echidna (EHH) and tiggywinkle (TWHH) hedgehog genes [45]. Our analyses show that the non-mammalian DHH orthologues do not cluster with the DHH of mammals, but instead form a highly divergent out-group, suggesting that these paralogues had an independent evolutionary origin (Figure 2; Additional file 8) and are hereafter referred to as the fishy hedgehogs (Fhh).
Phylogenetic tree showing divergence of the hedgehog family members in model organisms in which the genes have been completely sequenced. Mm = mouse, Me = tammar wallaby, Tt = dolphin, Gg = gorilla, Hs = human, Xt = Xenopus. Complete node labels and distances can be found in additional file 8. Each hedgehog subtype (IHH, SHH, DHH) forms a separate lineage. The fish DHH orthologues form a separate cluster from the mammalian DHH genes and have been renamed the fishy-hedgehog (FHH) cluster.
The PTCH analyses were less inclusive as only a few species had complete open reading frame sequences available. However, the PTCH1 phylogenetic tree mimicked standard mammalian groupings, with the tammar clustering with other mammals, and zebrafish as the outlier (Additional file 7b). Phylogenetic analysis of PTCH2 grouped tammar PTCH2 with that of other mammals, but as the most divergent lineage. PTCH2 was more divergent between species than DHH or PTCH1 (Additional file 7c).
All full-length proteins used for phylogenetic analyses were obtained from Ensembl and are listed in Additional file 2.
Genomic localisation of the tammar wallaby DHHcDNA
A single localization signal was observed for tammar wallaby DHH on chromosome 1q in approximately 50% of spreads examined, with no other consistent localization signals seen (Figure 3).
Expression of DHHin the tammar wallaby
DHH, PTCH1 and PTCH2 expression was examined in the gonads throughout fetal and pouch young development using an RT-PCR assay. In contrast to the mouse, DHH was expressed in the gonads of both male and female fetuses and pouch young. DHH was expressed from the first appearance of the genital ridge until after testicular and ovarian differentiation had occurred and persisted in the adult (Figure 4a). In addition, both PTCH1 and PTCH2 were expressed throughout gonadal development, through to adulthood. Although not quantitative, PTCH2 expression appeared to be expressed at a lower level than PTCH1. In the testis, both PTCH1 and PTCH2 were present from 1 day after birth, around the time of initiation of cord formation in the tammar (Figure 4a).
Expression of DHH, PTCH1 and PTCH2 in the tammar wallaby. a. RT-PCR for DHH, PTCH1 and PTCH2 in the tammar wallaby testis and ovary throughout development. DHH primers produce a band of 365 bp, PTCH1 primers produce a 130 bp product and PTCH2 primers produce a 330 bp product. d = day of fetal development from a 26.5 average gestation period. D0 = the day of birth. D = day of development post partum (pp). DHH expression was present at all time points during ovarian development. b-d. Quantitative PCR for DHH, PTCH1 and PTCH2. b. DHH was expressed at high levels in the developing gonads of both males and females (although it was lower in females overall). The lowest level of expression was observed in the adult ovary and the highest in the D4 male. Both PTCH1 (c) and PTCH2 (d) levels remained relatively consistent throughout gonad development. PTCH2 mRNA levels were always higher than PTCH1 in the gonads, and levels were consistently lowest for both genes in the adult ovary.
A limited quantitative real-time PCR profile confirmed the presence of DHH at all stages of gonad development in both testes and ovaries (Figure 4b-d). DHH mRNA levels were similar in the developing male and female gonads at all stages examined, but in adult gonads ovarian expression was low (Figure 4b). PTCH1 expression was consistent throughout gonadal development in males and females, with lowest levels seen in the adult ovary (Figure 4c). In contrast, PTCH2 levels remained constant in the gonad of both males and females at all stages examined including in the adult (Figure 4d). All three genes were also expressed in the prostate and adrenal albeit at lower levels than in the developing gonads.
DHH, PTCH1 and PTCH2 protein distribution during gonadogenesis
DHH was widely distributed throughout the bipotential gonad, PTCH1 and 2 staining was present but very weak (Additional file 9, d24 fetus). By day 1 pp the cords were beginning to form in the testis, and DHH stained pre-Sertoli cells that were coalescing into cord- like structures (Additional file 9, D1pp). PTCH1 protein stained weakly throughout the gonad while PTCH2 appeared more prominent and was localized outside the forming cords. At day 9 pp cords were fully formed in the developing testis. DHH protein was present throughout the gonad and was strongly detected in some Sertoli cells and in the peritubular myoid cells and at the basement membrane. PTCH1 was diffuse throughout the gonad but was mainly localised in the Sertoli cells and absent from the interstitium. Conversely, PTCH2 staining was more intense and concentrated in Leydig cells in the interstitium (Figure 5). In the adult testis, DHH was present at low levels in all cell types, but strong staining was seen in round spermatids from the post pachytene primary spermatocyte stage, through to the mature sperm. PTCH1 was present at high levels in the Leydig cells and showed a punctate distribution reminiscent of membrane bound protein recycling [46]. PTCH2 distribution also became highly restricted and localised strongly in the Sertoli cells (Figure 5).
Immunohistochemistry of DHH, PTCH1 and PTCH2 in the tammar wallaby testis at key developmental time points. Red/brown staining indicates protein distribution while the heamatoxalin counterstain appears blue. It is important to note that DHH is a highly secreted molecule and staining does not imply cell of origin. DHH staining was most intense at the basal lamina (BL). In the adult staining is concentrated in the develop spermatocytes (GC). At day D9pp PTCH1 was present within the Sertoli cells (SC) while PTCH2 was predominant in the Leydig cells (LC). This expression profile is reversed in the adult testis with PTCH1 found predominantly in the Leydig cells, while PTCH2 was predominate in the Sertoli cells. Scale bars indicate 40 μm, controls show immunohistochemistry with the primary antibody omitted.
The ovary becomes clearly differentiated around day 8 pp in the tammar [35]. At day 9 pp DHH staining was diffuse throughout the gonad along with PTCH1 and PTCH2 (Additional file 10). However, all three proteins were noticeably absent from the germ cells. By day 72 pp DHH was present in low levels across the ovary, but strongly localised within the germ cells that have coalesced into nests. PTCH1 was also expressed in the germ cells at this stage, while PTCH2 staining was weak and primarily in the interstitial tissue (Additional file 9). In the adult ovary, DHH was present but weak throughout the gonad. PTCH1 staining was only weakly detected in the granulosa cells of antral follicles, but increased in the cumulus cells of mature follicles. Staining was also seen in the steroidogenic theca cells. PTCH2 showed a similar distribution to PTCH1 but was abundant in the granulosa, cumulus and theca cells (Figure 6). DHH, PTCH1, and PTCH2 were also detected in the corpus luteum (Additional file 9).
Immunohistochemistry of DHH, PTCH1 and PTCH2 in the tammar wallaby ovary. DHH was found in the granulosa cells (GC) of follicles at all stages of development, and in the oocyte cytoplasm shown in primordial follicles (PrF). Staining was also observed for all three proteins in the cumulus cells (CC). PTCH1 and PTCH2 showed a similar distribution in the granulosa cells and were also present at reduced levels in the theca (T). PTCH2 staining was evident in the oocyte but PTCH1 was not. Scale bars indicate 160 μm, controls show immunohistochemistry with the primary antibody omitted.
Discussion
We have shown that DHH is a highly conserved mammal specific hedgehog paralogue with conserved expression during mammalian gonadogenesis. DHH and its receptors PTCH1 and 2 are highly conserved at the protein level and are expressed in an analogous pattern to that seen in the mouse gonad. However, DHH was expressed in the developing marsupial ovary in contrast to the mouse, in which it is testis-specific during development.
Phylogenetic analysis of the hedgehog gene family across vertebrates shows that non-mammalian DHH genes in fish form a distinct subgroup, distantly related to mammalian DHH genes, indicating they have had an independent evolutionary origin. We have re-named this sub-group fishy hedgehog (FHH) to emphasise their distinction from the DHH genes. This suggests that the evolution of mammalian DHH is a recent event (Figure 2) making it quite unique among the gonadal differentiation genes, all of which have orthologues in the non-mammalian vertebrates with the notable exception of the sex determination switch gene SRY, which is also mammal specific [47]. Despite its recent origin, DHH was extremely highly conserved between marsupials and eutherians, suggesting it quickly adopted an essential function in mammalian reproduction.
The hedgehog receptors PTCH1 and 2 were highly conserved between marsupials, eutherians and non-mammalian vertebrates. Marsupial PTCH2 was the most divergent (especially in the C-terminal region, consistent with findings in other vertebrate species [48]) but still shared 89% amino acid similarity with eutherian orthologues. The tammar PTCH2 C-terminus contained a 70 amino acid additional exon not found in eutherian PTCH2 proteins. Interestingly, significant homology to the additional tammar exon was identified in the human PTCH2 genomic sequence, in intron 21, and shared 70% identity at the nucleotide level and 79% amino acid similarity with the tammar additional exon (hereafter referred to as exon 21a). The level of conservation of this exon between marsupials and humans was much higher than that of non-functional intronic DNA, suggesting functional conservation of the sequence. Translation of the human sequence revealed a premature stop codon at amino acid 35, so its inclusion in the transcript would lead to a PTCH2 receptor with a severely truncated intracellular signalling domain (Figure 1b). Such an isoform, lacking exon 22, identical to the one predicted from the inclusion of the human putative exon 21a, has been previously identified (the Δ-22 isoform)[18]. The human Δ-22 PTCH2 isoform is the only one capable of acting as a strong inhibitor of SHH induction, similar in function to PTCH1 [18]. It appears that the ability to produce such an isoform was derived from a stochastic nonsense mutation in the original exon 21a leading to a truncated protein. The tammar does not have a premature stop (exon 21a is an intact ORF) and so this tammar PTCH2 isoform does not share redundancy with PTCH1 function. The degree of conservation of this region in humans suggests that it has only recently become non-functional in primate evolution. It is intriguing then, that this sequence could not be identified in any other mammalian PTCH2 loci, but only in the tammar, opossum, and human. These findings suggest that the exon was present in the ancestral PTCH2 gene and has been independently lost in different eutherian lineages (Figure 1c). We also identified several PTCH2 isoforms that appear to be dynamically regulated at specific developmental time points. This is also consistent with findings in humans that identified PTCH2 isoforms lacking exons 9 and 10 (PTCH2-Δ9-10)[18]. Taken together, these data suggest that PTCH2 has divergent species-specific roles in development, while PTCH1 is likely to maintain a highly conserved function in hedgehog signal transduction. Furthermore, it suggests that the human PTCH2 Δ-22 isoform may have evolved to compensate for a loss of PTCH1 in tissues in which they are co-expressed.
DHH, PTCH1 and PTCH2 mRNA and protein were present throughout gonadal development in both males and females, from early development through to adult stages. The presence of ligand and both receptor proteins throughout gonadal development is consistent with findings in mouse testis, but not ovary [49] and suggests a conserved role for hedgehog signalling in mammalian gonad formation. These findings are also consistent with the observed disruption to normal gonadal patterning and significant reduction in the expression of the downstream target gene GLI1, in the tammar when hedgehog signalling is ablated in vitro [37].
In the testis, DHH could be seen within the pre-Sertoli cells of the aggregating cords. Once the testes differentiated, DHH staining was concentrated in the Sertoli cells, especially at the basal lamina of the cord. This protein distribution is similar to that reported in mouse [49, 24, 49] and suggests it is critical for testicular patterning. However, there were some differences in PTCH distribution from predicted mouse patterns. PTCH1 staining was similar to that of DHH, and was distributed mainly within the seminiferous cords (containing germ cells and Sertoli cells). This was in contrast to the interstitial expression seen for Ptch1 in the developing mouse testis [24] but similar to the expression of Ptch2 [17]. Conversely, PTCH2 staining in the tammar was more reminiscent of Ptch1 distribution in the mouse testis [17] and was located throughout the gonad but concentrated in the interstitium and Leydig cells. This suggests there may have been a reversal in the roles of these receptors in marsupial testicular development relative to the mouse. Since detailed localisation of the PTCH receptors during gonad development in other mammals and vertebrates is not available we cannot determine which profile is more typical during development.
There was discrete staining of DHH, PTCH1 and PTCH2 proteins in the adult testis. DHH was concentrated in the differentiating germ cells, but restricted to the post pachytene primary spermatocyte stage through to the mature sperm. There was faint PTCH1 and PTCH2 staining throughout the testis but the proteins were concentrated in the Leydig cells and Sertoli cells respectively. This is consistent with in situ results in the adult mouse testis [17], suggesting a conserved role for these genes in maintaining testicular function and spermatogenesis in all therian mammals.
Unlike in the mouse, in which Dhh is testis-specific in early development [1], DHH was expressed in the developing tammar ovary throughout development. Activation of hedgehog signalling in the developing mouse ovary leads to Leydig cell development [25]. However, early ovarian development was not affected by the presence of DHH in the tammar, despite the presence of similar mRNA and protein levels of both ligand and receptors as in the developing testis. These findings show that SRY is not needed for DHH activation in the developing gonad. In the juvenile ovary, DHH was abundant in the oocytes consistent with the suggested role for DHH in maintaining the germ line [1]. In the adult ovary, DHH was broadly co-localized with PTCH1 and 2, in follicles and the corpus luteum suggesting it may be important for normal folliculogenesis and steroidogenesis, consistent with recent findings in the mouse [26]. As in the testis, PTCH2 appeared to be the predominant receptor throughout ovarian development and in the adult.
Conclusions
These data support a conserved role for hedgehog signalling in gonadal development but show that in marsupials this pathway may be significant for early patterning of the ovary as well as for the testis. These results, in conjunction with our phylogenetic analysis of hedgehog family members in all vertebrates, suggests that DHH is unique to mammals and is a conserved member of the gonadal development pathway.
References
Bitgood MJ, Shen L, McMahon AP: Sertoli cell signaling by Desert hedgehog regulates the male germline. Curr Biol. 1996, 6: 298-304. 10.1016/S0960-9822(02)00480-3.
Kamisago M, Kimura M, Furutani Y, Furutani M, Takao A, Momma K, Matsuoka R: Assignment of human desert hedgehog gene (DHH) to chromosome band 12q13.1 by in situ hybridization. Cytogenet Cell Genet. 1999, 87: 117-118. 10.1159/000015376.
Kumar S, Balczarek KA, Lai ZC: Evolution of the hedgehog gene family. Genetics. 1996, 142: 965-972.
Pathi S, Pagan-Westphal S, Baker DP, Garber EA, Rayhorn P, Bumcrot D, Tabin CJ, Blake Pepinsky R, Williams KP: Comparative biological responses to human Sonic, Indian, and Desert hedgehog. Mech Dev. 2001, 106: 107-117. 10.1016/S0925-4773(01)00427-0.
Tate G, Satoh H, Endo Y, Mitsuya T: Assignment of Desert Hedgehog (DHH) to human chromosome bands 12q12 - > q13.1 by in situ hybridization. Cytogenetics And Cell Genetics. 2000, 88: 93-94. 10.1159/000015495.
Ingham PW: Developmental biology - Boning up on Hedgehog's movements. Nature. 1998, 394: 16-17. 10.1038/27764.
Mirsky R, Jessen KR: The neurobiology of Schwann cells. Brain Pathol. 1999, 9: 293-311.
Patten I, Placzek M: The role of Sonic hedgehog in neural tube patterning. Cellular And Molecular Life Sciences. 2000, 57: 1695-1708. 10.1007/PL00000652.
Perron M, Boy S, Amato MA, Viczian A, Koebernick K, Pieler T, Harris WA: A novel function for Hedgehog signalling in retinal pigment epithelium differentiation. Development. 2003, 130: 1565-1577. 10.1242/dev.00391.
Echelard Y, Epstein DJ, Stjacques B, Shen L, Mohler J, McMahon JA, McMahon AP: Sonic-Hedgehog, A Member Of A Family Of Putative Signaling Molecules, Is Implicated In The Regulation Of Cns Polarity. Cell. 1993, 75: 1417-1430. 10.1016/0092-8674(93)90627-3.
van den Brink GR, Bleuming SA, Hardwick JC, Schepman BL, Offerhaus GJ, Keller JJ, Nielsen C, Gaffield W, van Deventer SJ, Roberts DJ, Peppelenbosch MP: Indian Hedgehog is an antagonist of Wnt signaling in colonic epithelial cell differentiation. Nat Genet. 2004, 36: 277-282. 10.1038/ng1304.
Spicer LJ, Sudo S, Aad PY, Wang LS, Chun SY, Ben-Shlomo I, Klein C, Hsueh AJ: The hedgehog-patched signaling pathway and function in the mammalian ovary: a novel role for hedgehog proteins in stimulating proliferation and steroidogenesis of theca cells. Reproduction. 2009, 138: 329-339. 10.1530/REP-08-0317.
Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ: Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996, 273: 613-622. 10.1126/science.273.5275.613.
Szczepny A, Hime GR, Loveland KL: Expression of hedgehog signalling components in adult mouse testis. Dev Dyn. 2006, 235: 3063-3070. 10.1002/dvdy.20931.
Mirsky R, Parmantier E, McMahon AP, Jessen KR: Schwann cell-derived desert hedgehog signals nerve sheath formation. Ann N Y Acad Sci. 1999, 883: 196-202. 10.1111/j.1749-6632.1999.tb08582.x.
Parmantier E, Lynn B, Lawson D, Turmaine M, Namini SS, Chakrabarti L, McMahon AP, Jessen KR, Mirsky R: Schwann cell-derived Desert hedgehog controls the development of peripheral nerve sheaths. Neuron. 1999, 23: 713-724. 10.1016/S0896-6273(01)80030-1.
Carpenter D, Stone DM, Brush J, Ryan A, Armanini M, Frantz G, Rosenthal A, de Sauvage FJ: Characterization of two patched receptors for the vertebrate hedgehog protein family. Proc Natl Acad Sci USA. 1998, 95: 13630-13634. 10.1073/pnas.95.23.13630.
Rahnama F, Toftgard R, Zaphiropoulos PG: Distinct roles of PTCH2 splice variants in Hedgehog signalling. Biochem J. 2004, 378: 325-334. 10.1042/BJ20031200.
Nusslein-Volhard C, Wieschause E: Mutation affecting segment number and polarity in Drosophila. Nature (london). 1980, 287: 795-801. 10.1038/287795a0.
Bitgood MJ, McMahon AP: Hedgehog And Bmp Genes Are Coexpressed At Many Diverse Sites Of Cell-Cell Interaction In The Mouse Embryo. Developmental Biology. 1995, 172: 126-138. 10.1006/dbio.1995.0010.
Bitgood MJ, Shen LY, McMahon AP: Sertoli cell signaling by Desert hedgehog regulates the male germline. Current Biology. 1996, 6: 298-304. 10.1016/S0960-9822(02)00480-3.
Sekido R, Lovell-Badge R: Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature. 2008, 453: 930-934. 10.1038/nature06944.
Clark AM, Garland KK, Russell LD: Desert hedgehog (Dhh) gene is required in the mouse testis for formation of adult-type Leydig cells and normal development of peritubular cells and seminiferous tubules. Biology Of Reproduction. 2000, 63: 1825-1838. 10.1095/biolreprod63.6.1825.
Yao HH, Whoriskey W, Capel B: Desert Hedgehog/Patched 1 signaling specifies fetal Leydig cell fate in testis organogenesis. Genes Dev. 2002, 16: 1433-1440. 10.1101/gad.981202.
Barsoum IB, Bingham NC, Parker KL, Jorgensen JS, Yao HH: Activation of the Hedgehog pathway in the mouse fetal ovary leads to ectopic appearance of fetal Leydig cells and female pseudohermaphroditism. Dev Biol. 2009, 329: 96-103. 10.1016/j.ydbio.2009.02.025.
Russell MC, Cowan RG, Harman RM, Walker AL, Quirk SM: The hedgehog signaling pathway in the mouse ovary. Biol Reprod. 2007, 77: 226-236. 10.1095/biolreprod.106.053629.
Umehara F, Tate G, Itoh K, Yamaguchi N, Douchi T, Mitsuya T, Osame M: A novel mutation of desert hedgehog in a patient with 46, XY partial gonadal dysgenesis accompanied by minifascicular neuropathy. Am J Hum Genet. 2000, 67: 1302-1305.
Canto P, Soderlund D, Reyes E, Mendez JP: Mutations in the Desert hedgehog (DHH) gene in patients with 46, XY complete pure gonadal dysgenesis (vol 89, pg 4480, 2004). Journal Of Clinical Endocrinology And Metabolism. 2004, 89: 5453-5453.
Bagheri-Fam S, Sinclair AH, Koopman P, Harley VR: Conserved regulatory modules in the Sox9 testis-specific enhancer predict roles for SOX, TCF/LEF, Forkhead, DMRT, and GATA proteins in vertebrate sex determination. Int J Biochem Cell Biol. 2010, 42: 472-477. 10.1016/j.biocel.2009.07.001.
Bininda-Emonds OR, Cardillo M, Jones KE, MacPhee RD, Beck RM, Grenyer R, Price SA, Vos RA, Gittleman JL, Purvis A: The delayed rise of present-day mammals. Nature. 2007, 446: 507-512. 10.1038/nature05634.
Renfree MB, Tyndale-Biscoe CH: Intrauterine development after diapause in the marsupial Macropus eugenii. Dev Biol. 1973, 32: 28-40. 10.1016/0012-1606(73)90217-0.
Tyndale-Biscoe CH, Hearn JP, Renfree MB: Control of reproduction in macropodid marsupials. J Endocrinol. 1974, 63: 589-614. 10.1677/joe.0.0630589.
O WS, Short RV, Renfree MB, Shaw G: Primary genetic control of somatic sexual differentiation in a mammal. Nature. 1988, 331: 716-717. 10.1038/331716a0.
Harry JL, Koopman P, Brennan FE, Graves JA, Renfree MB: Widespread expression of the testis-determining gene SRY in a marsupial. Nat Genet. 1995, 11: 347-349. 10.1038/ng1195-347.
Renfree MB, O WS, Short RV, Shaw G: Sexual differentiation of the urogenital system of the fetal and neonatal tammar wallaby, Macropus eugenii. Anat Embryol (Berl). 1996, 194: 111-134.
Church GM, Gilbert W: Genomic sequencing. Proc Natl Acad Sci USA. 1984, 81: 1991-1995. 10.1073/pnas.81.7.1991.
Chung JW, Pask AJ, Renfree MB: Seminiferous cord formation is regulated by Hedgehog signalling in the marsupial. Reproductive Biology. 2011
Renfree MB, Papenfuss AT, Deakin JE, Lindsay J, Heider T, Belov K, Rens W, Waters PD, Pharo EA, Shaw G, et al: Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development. Genome Biol. 2011, 12: R81-10.1186/gb-2011-12-8-r81.
Schempp W, Meer B: Cytologic evidence for three human X-chromosomal segments escaping inactivation. Hum Genet. 1983, 63: 171-174. 10.1007/BF00291539.
Wilcox SA, Watson JM, Spencer JA, Graves JA: Comparative mapping identifies the fusion point of an ancient mammalian X-autosomal rearrangement. Genomics. 1996, 35: 66-70. 10.1006/geno.1996.0323.
Pask AJ, Calatayud NE, Shaw G, Wood WG, Renfree MB: Estrogen blocks the nuclear entry of SOX9 in the developing gonad of a marsupial mammal. BMC Biology. 2010
Pfaffl MW: A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29: e45-10.1093/nar/29.9.e45.
Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, McMahon JA, McMahon AP: Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993, 75: 1417-1430. 10.1016/0092-8674(93)90627-3.
Porter JA, Ekker SC, Park WJ, von Kessler DP, Young KE, Chen CH, Ma Y, Woods AS, Cotter RJ, Koonin EV, Beachy PA: Hedgehog patterning activity: role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell. 1996, 86: 21-34. 10.1016/S0092-8674(00)80074-4.
Lewis KE, Eisen JS: Hedgehog signaling is required for primary motoneuron induction in zebrafish. Development. 2001, 128: 3485-3495.
Musinova YR, Lisitsyna OM, Golyshev SA, Tuzhikov AI, Polyakov VY, Sheval EV: Nucleolar localization/retention signal is responsible for transient accumulation of histone H2B in the nucleolus through electrostatic interactions. Biochim Biophys Acta. 2011, 1813: 27-38. 10.1016/j.bbamcr.2010.11.003.
Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN: A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990, 346: 240-244. 10.1038/346240a0.
Motoyama J, Takabatake T, Takeshima K, Hui C: Ptch2, a second mouse Patched gene is co-expressed with Sonic hedgehog. Nat Genet. 1998, 18: 104-106. 10.1038/ng0298-104.
Goodrich LV, Johnson RL, Milenkovic L, McMahon JA, Scott MP: Conservation of the hedgehog/patched signaling pathway from flies to mice: induction of a mouse patched gene by Hedgehog. Genes Dev. 1996, 10: 301-312. 10.1101/gad.10.3.301.
Acknowledgements
We thank Drs Chai-an Mao, Jenny L. Harry and Deanne J. Whitworth for help with the initial cloning of the DHH gene in the tammar wallaby. We also thank Kerry Martin and Scott Brownlees for assistance with the animals. This study was supported by The University of Connecticut Faculty Large Grant, a National Health and Medical Research Council project grant to MBR and AJP, an R D Wright Fellowship to AJP, the Australian Research Council Centre of Excellence in Kangaroo Genomics, a Federation Fellowship to MBR and an NIH research grant (HD30284) to RRB. There is no financial or other potential conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
All authors participated in the design of the study. Tissues were collected by WJA, AJP and MBR. Experiments were performed by WAO'H, WJA and AJP. All authors analyzed the results. AJP, WJA, WAO'H and MBR drafted the manuscript. All authors read, modified and approved the final manuscript.
William A O'Hara, Walid J Azar contributed equally to this work.
Electronic supplementary material
12861_2011_679_MOESM3_ESM.PDF
Additional file 3: a. Schematic diagram of the alternative splice variants detected for tammar wallaby PTCH2 relative to the human PTCH2 structure. Primers spanned exons 16-22 (red bar) and 7 splice variants (including the full length transcript) were isolated. Tammar PTCH2 has two additional introns in exon 17 and 22 (blue arrow heads) and one additional exon (21-Me; red arrow). b. Table showing the relative homologies of the epitope to which the DHH antibody was raised (recombinant mouse (Rm) Dhh amino acids 199-396) to tammar wallaby DHH, SHH and IHH. Homology is significantly lower with SHH and IHH. c. Western Blot of DHH antibody a band at 43 kDa, which is the predicted size of the tammar wallaby DHH protein in its uncleaved form. Antibody cross-reactivity with SHH or IHH would create bands at 48 and 45 kDa respectively. (PDF 786 KB)
12861_2011_679_MOESM4_ESM.PDF
Additional file 4: Alignment of tammar Dhh protein sequence with four eutherian mammals. Dark shading indicates agreement in at least 60% of the sequences, light shading indicates amino acid similarity to consensus. Double dashed area represents conserved sequence necessary for secreted DHH. Asterisked region represents conserved catalytic site. (PDF 33 KB)
12861_2011_679_MOESM5_ESM.PDF
Additional file 5: Alignment of tammar Ptch1 protein sequence with four eutherian mammals. Dark shading indicates agreement in at least 60% of the sequences, light shading indicates amino acid similarity to consensus. Double dashed areas represent putative trans-membrane binding domains, with species (Ptch1 unless indicated) showing highest sequence identity indicated in parentheses. Any conserved domains are mentioned above the relative sequence. Glycosylation sites are denoted with a cross. (PDF 67 KB)
12861_2011_679_MOESM6_ESM.PDF
Additional file 6: a. Alignment of tammar Ptch2 protein sequence with four eutherian mammals. Dark shading indicates agreement in at least 60% of the sequences, light shading indicates amino acid similarity to consensus. 70 amino acid stretch maintained in Tammar is italicized. Double dashed areas represent putative trans-membrane binding domains, with species showing highest sequence identity indicated in parentheses. Any conserved domains are mentioned above the relative sequence. b. Alternative splice variants of PTCH2. Primers were designed to span the region corresponding to exons 18-22 of the human PTCH2 gene. RT-PCR was carried out in day 3, 7 and 14 post partum testes. Day 3 PCR produced four bands of ~1.2 Kb, 1.05 Kb, 950 bp and 860 bp. We sequence verified that the 1.2 Kb fragment was the full-length transcript and that the 950 bp transcript was a Δ-21a PTCH2 isoform. The identity of the missing exons in the 1.05 Kb and 860 bp fragments is shown in Additional File 9. These slice variants were developmentally regulated, with the smaller two isoforms not seen in the day 7 or 14 testis and the larger two isoforms appear to change in their relative abundance between stages. (PDF 173 KB)
12861_2011_679_MOESM7_ESM.PDF
Additional file 7: a-c. Phylogenetic trees showing divergence of DHH(A), PTCH1(B), and PTCH2(C) in model organisms in which the genes have been completely sequenced. Mm = mouse, La = elephant, Me = tammar wallaby, Cf = dog, Tt = dolphin, Ss = pig, Gg = gorilla, Hs = human, Pp = Chimpanzee, Bt = cow, Dr = zebrafish, Tn = Tetraodon, Ol = Oryzias. Zebrafish is included in b and c as a known outlier. (PDF 124 KB)
12861_2011_679_MOESM8_ESM.PDF
Additional file 8: Phylogenetic tree showing clustering of all complete sequenced HH proteins (Indian (IHH), Sonic (SHH), desert (DHH), Echidna (EHH), TwiggyWinkle (TWHH)). EHH and TWHH each contain only 1 member, and have both been shown to cluster within IHH and SHH groups respectively. The fish DHH orthologues (FHH) form a separate cluster from the mammalian DHH genes. EHH, TWHH and reported fish DHH orthologues (FHH) are highlighted in red. Node labels are in the format: PROTEIN_Genus_species. (PDF 177 KB)
12861_2011_679_MOESM9_ESM.PDF
Additional file 9: Immunohistochemistry of DHH, PTCH1 and PTCH2 in the tammar wallaby testis at key developmental time points. Red/brown staining indicates protein distribution while the heamatoxalin counterstain appears blue. It is important to note that DHH is a highly secreted molecule and staining does not imply cell of origin. DHH was initially present at high levels throughout the indifferent gonad (d24 fetus), by D1pp Dhh is confined to the aggregating seminiferous cords (AC). Scale bars = 36 μm. (PDF 14 MB)
12861_2011_679_MOESM10_ESM.PDF
Additional file 10: Immunohistochemistry of DHH, PTCH1 and PTCH2 in the tammar wallaby ovary at key developmental time points. At day 9pp when the ovary is forming a cortex and medulla, there was widespread staining for DHH, PTCH1 and PTCH2 throughout the ovary. By D72pp DHH and PTCH1 were concentrated in the germ cell nests (CGN) and PTCH2 was largely in the interstitium. In the adult ovary, DHH was found in the granulosa cells (GC) of follicles at all stages of development, and in the oocyte cytoplasm. Staining was also observed in the corpus luteum (CL). Scale bars = 40 μm at D9, D72 and 160 μm in the corpus luteum. (PDF 8 MB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
O'Hara, W.A., Azar, W.J., Behringer, R.R. et al. Desert hedgehogis a mammal-specific gene expressed during testicular and ovarian development in a marsupial. BMC Dev Biol 11, 72 (2011). https://doi.org/10.1186/1471-213X-11-72
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-213X-11-72