Abstract
Background
In yeast, glucose-dependent degradation of the Mth1 protein, a corepressor of the glucose transporter gene (HXT) repressor Rgt1, is a crucial event enabling expression of several HXT. This event occurs through a signaling pathway that involves the Rgt2 and Snf3 glucose sensors and yeast casein kinase 1 and 2 (Yck1/2). In this study, we examined whether the glucose sensors directly couple with Yck1/2 to convert glucose binding into an intracellular signal that leads to the degradation of Mth1.
Results
High levels of glucose induce degradation of Mth1 through the Rgt2/Snf3 glucose signaling pathway. Fluorescence microscopy analysis indicates that, under glucose-limited conditions, GFP-Mth1 is localized in the nucleus and does not shuttle between the nucleus and cytoplasm. If glucose-induced degradation is prevented due to disruption of the Rgt2/Snf3 pathway, GFP-Mth1 accumulates in the nucleus. When engineered to be localized to the cytoplasm, GFP-Mth1 is degraded regardless of the presence of glucose or the glucose sensors. In addition, removal of Grr1 from the nucleus prevents degradation of GFP-Mth1. These results suggest that glucose-induced, glucose sensor-dependent Mth1 degradation occurs in the nucleus. We also show that, like Yck2, Yck1 is localized to the plasma membrane via C-terminal palmitoylation mediated by the palmitoyl transferase Akr1. However, glucose-dependent degradation of Mth1 is not impaired in the absence of Akr1, suggesting that a direct interaction between the glucose sensors and Yck1/2 is not required for Mth1 degradation.
Conclusion
Glucose-induced, glucose sensor-regulated degradation of Mth1 occurs in the nucleus and does not require direct interaction of the glucose sensors with Yck1/2.
Similar content being viewed by others
Background
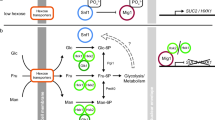
In the budding yeast Saccharomyces cerevisiae, glucose stimulates its transport across the plasma membrane by inducing expression of several HXT[1–3]. Under glucose-limited conditions, the transcriptional repressor Rgt1 binds to the HXT promoters and recruits general corepressors Ssn6 and Tup1 [4–7]. Rgt1 does this in conjunction with its corepressor Mth1, which physically interacts with Rgt1 [8–10]. Therefore, it has been proposed that Rgt1 forms a repression complex with Mth1, Ssn6, and Tup1 on the HXT promoters, inhibiting transcription [6]. Glucose appears to prevent formation of this protein complex by causing degradation of Mth1, resulting in release of Rgt1 from HXT promoters, thereby inducing expression of HXT[6, 11–14].
The glucose signal that leads to degradation of Mth1 is generated by the plasma-membrane spanning glucose sensors Rgt2 and Snf3. Signal generation is a receptor-mediated process and does not require glucose metabolism. This idea is supported by evidence that dominant mutations exist in the glucose sensor genes that lock the sensor proteins in glucose-bound conformations, generating a constitutive signal [15, 16]. Indeed, Mth1 is constitutively degraded in cells expressing the active glucose sensor mutants [17]. Subsequent studies have shown that the plasma membrane-tethered casein kinases Yck1/2 phosphorylate Mth1, triggering its ubiquitination and subsequent degradation [18]. It has also been shown by yeast-two-hybrid assay that Mth1 interacts with the C-terminal tails of the glucose sensors, suggesting that Mth1 is recruited to the plasma membrane [19–21]. These observations have led to the current view of glucose-induced HXT expression. Upon glucose binding, the glucose sensors are converted from inactive to active forms through a conformational change, activating Yck1/2 in their vicinity. Mth1, recruited by the glucose sensors to the plasma membrane, is phosphorylated by Yck1/2 and, subsequently, ubiquitinated by SCFGrr1. Finally, the ubiquitinated Mth1 is targeted for degradation by the 26S proteasome [12–14, 17].
However, this hypothesis is mainly based on the following assumptions: (1) Mth1 is excluded from the nucleus upon glucose addition and recruited to the plasma membrane by any means, and (2) Yck1/2 are activated through a direct interaction with the glucose sensors. In this study, we specifically tested these assumptions and provide evidence that Yck1/2 do not directly couple to the glucose sensors during transmission of the glucose signal from the plasma membrane to the nucleus. A possible mechanism for how the glucose sensors and Yck1/2 collaborate to degrade Mth1 is discussed.
Results
Subcellular localization of Mth1 is not regulated
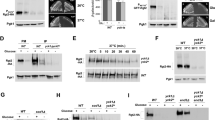
We have previously reported that GFP-Mth1 is mainly nuclear in glucose-depleted medium and is rapidly degraded upon glucose addition [14]. To gain more insights into glucose-dependent degradation of Mth1, we first examined subcellular localization of GFP-Mth1 in cells grown under conditions where HXT expression is repressed (2% galactose) or induced (4% glucose) (Figure 1). Because glucose also regulates expression of MTH1[22], GFP-MTH1 was expressed from the MET25 promoter, which is not regulated by glucose, in a low copy centromeric plasmid [14]. Fluorescence microscopy analysis demonstrates that GFP-Mth1 is localized primarily to two specific foci within the nucleus and that ~50% of fluorescence is recovered within 30-40 seconds after photobleaching (Figure 1A). These results suggest that, under glucose-limited conditions, Mth1 predominantly accumulates in the nucleus but does not shuttle between the nucleus and cytoplasm. We also monitored glucose-induced degradation of Mth1 in a time-lapse manner and observed that more than half of GFP-Mth1 is degraded within 5-10 min after glucose addition (Figure 1B), which is consistent with the Western blot analysis obtained previously [14].
Mth1 does not shuttle between the nucleus and cytoplasm. (A) Images of GFP-Mth1 expressing yeast cells grown in galactose (2%) before and after photobleaching in the indicated region. One of two foci (arrow) was bleached and then recovery of the fluorescence in the bleached zone was monitored. Time is in seconds after photobleaching. (B) Time-lapse observation of Mth1 degradation in high-glucose medium (4%). Numbers indicate time in minutes. Yeast cells, wild-type (WT; BY4742) and grr1 mutant (grr1Δ; YM4783), expressing GFP-Mth1 were observed under the Zeiss LSM 510 META Confocal Laser Scanning Microscope (CLSM).
To determine whether Mth1 is excluded from the nucleus in response to glucose, we determined in which cellular compartment Mth1 is localized when expressed under high glucose conditions. Toward this aim, we examined subcellular localization of GFP-Mth1 in the rgt2snf3 and grr1 mutants, where glucose-dependent degradation of Mth1 is severely impaired [14]. As shown in Figure 2A, the GFP-Mth1 proteins accumulate in the nucleus of the mutants grown in high-glucose medium. To further test a possibility that the glucose sensors regulate nuclear exclusion of Mth1, we determined subcellular localization of a nondegradable form of Mth1 in wild-type cells, where the glucose sensors are active. The dominant HTR1-23 mutation in MTH1 (I85S) [23] converts Mth1 into a degradation-resistant form [14]. Our results indicate that the mutant Mth1 proteins (GFP-Mth1-I85S) are not degraded, as expected, but accumulate in the nucleus in high-glucose medium (Figure 2B). We also observed that subcellular localization and degradation of Mth1 are not influenced by the absence of Rgt1, a downstream target of the Rgt2/Snf3-mediated signaling pathway [22]. Therefore, it is likely that neither glucose nor the glucose sensors control nuclear export of Mth1.
Neither glucose nor the glucose sensors influence the subcellular localization of Mth1. (A) Yeast cells of the indicated genotype expressing GFP-Mth1 were grown to mid-log phase in a selective medium containing 2% galactose. Aliquots were then transferred to 2% galactose medium (Gal) or 4% glucose medium (Glu) and incubated for 60 min. Subcellular localization of GFP-Mth1 was analyzed by fluorescence microscopy. Yeast cells used were: WT (BY4742), rgt2Δsnf3Δ (YM6370), grr1Δ (YM4783), and rgt1Δ (YM4509). (B) Localization of a nondegradable form of Mth1 (Mth1-I85S) in wild-type cells (BY4742). (C) GFP-Rgt1 localization as a positive control.
Glucose-dependent degradation of Mth1 occurs in the nucleus
To more directly assess whether Mth1 degradation occurs in the nucleus or cytoplasm, we examined degradation of GFP-Mth1 proteins engineered to have cytoplasmic localization. To do so, GFP-Mth1 was tagged with the nuclear export signal (NES) of the yeast PKIα (GFP-NES-Mth1) [24]. To test the functionality of the NES motif, we constructed plasmid expressing GFP-NES(m)-Mth1 containing a single amino acid substitution within the motif. The levels and subcellular localization of the resulting fusion proteins were determined by Western blot and fluorescence microscopy analysis, respectively (Figure 3). We observed that GFP-NES-Mth1 is excluded from the nucleus, as expected (Figure 3A, bottom panel), but significantly degraded regardless of the presence of glucose or the glucose sensors (Figure 3B). In contrast, GFP-Mth1 with a nonfunctional NES motif (GFP-NES(m)-Mth1) behaves like GFP-Mth1; it accumulates in the nucleus in the absence of glucose (2% Gal) but is degraded when glucose is present (4% Glu), providing evidence that the NES motif used is functional. Therefore, collectively, these results suggest that glucose-dependent degradation of Mth1 occurs in the nucleus.
Glucose-dependent degradation of Mth1 takes place in the nucleus. Synthetic oligonucleotides encoding the nuclear export sequence (NES) and its mutant peptide (NES (m)) were inserted between the GFP and MTH1 genes in the GFP-MTH1 construct. Yeast cells (mth1Δ, YM6266) expressing GFP-Mth1 and its derivatives were grown as described in Figure 2. Subcellular localization and levels of GFP-Mth1 were analyzed by fluorescence microscopy using CLSM (A) and Western blotting using anti-GFP antibody (B), respectively.
Nuclear degradation of Mth1 is Grr1-dependent
Grr1 appears to be present in both the nucleus and cytoplasm, but deletion of the putative NLS in the amino-terminus of Grr1 (Grr1ΔN) causes it to be localized to the cytoplasm [25]. Previous studies have shown that expression of Grr1ΔN is sufficient to restore the morphology of, and to mediate degradation of Gic2 in, the grr1 mutant [25, 26], suggesting that Grr1ΔN is fully functional in the cytoplasm. To provide more compelling evidence that Grr1-dependent degradation of Mth1 takes place in the nucleus, we examined if GFP-Grr1ΔN (Δ1 280) can mediate Mth1 degradation (Figure 4). Our results show that GFP-Grr1ΔN is predominantly localized to the cytoplasm (Figure 4A) and able to restore the morphology of the grr1 mutant (Figure 4B), as reported previously [25, 26]. However, expression of Grr1ΔN in the grr1 mutant does not cause degradation of GFP-Mth1 (Figure 4C), probably due to translocation of Grr1ΔN from the nucleus to the cytoplasm (Figure 4B). These results suggest that SCFGrr1-mediated degradation of Mth1 occurs in the nucleus, not in the cytoplasm.
Glucose-dependent degradation of Mth1 does not occur when Grr1 is removed from the nucleus. (A) GFP-Grr1 and GFP-Grr1ΔN (lacking the first 280 amino acids of Grr1) were expressed in the grr1Δ mutant (YM4783) in high-glucose medium (4%). (B) GFP-Mth1 was expressed alone (top) or coexpressed with Grr1-ΔN (bottom) in the grr1 mutant (YM4783). Subcellular localization of GFP-Mth1 and GFP-Grr1ΔN was analyzed by fluorescence microscopy (A and B). (C) GFP-Mth1 was expressed alone (top) or coexpressed with Grr1-ΔN (bottom) in the grr1 mutant (YM4783), and levels of GFP-Mth1 were analyzed by Western blotting using anti-GFP antibody. Yeast cells were grown in 2% galactose medium (Gal) or 4% glucose medium (Glu) as described in Figure 2.
Direct interaction between Yck1/2 and the glucose sensors is not required for glucose-dependent degradation of Mth1
Next, we tested whether the glucose sensors are coupled with Yck1/2 in order to transduce glucose binding into an intracellular signaling cascade that leads to Mth1 degradation. Toward this aim, we examined whether Mth1 is degraded when Yck1/2 are mislocalized from the plasma membrane. It has been well described that Yck2 is targeted to the plasma membrane through palmitoylation of the C-terminal Cys-Cys sequence by the palmitoyl transferase Akr1 [27–29]. Confocal microscopy images show that, like GFP-Yck2 [29], GFP-Yck1 is associated with the plasma membrane in wild-type cells, but diffused throughout the entire cell when Akr1 is absent (Figure 5A, akr1Δ). Importantly, glucose-induced degradation of Mth1 is not significantly impaired in the absence of Akr1 (Figure 5B). These results suggest that neither localization of Yck1/2 to the plasma membrane nor a direct interaction between Yck1/2 and the glucose sensors is required for degradation of Mth1.
Direct interaction of Yck1/2 with the glucose sensors is not required for degradation of Mth1. (A) Subcellular localization of GFP-Yck1 and GFP-Yck2 expressed in wild type (BY4742) and akr1Δ mutant (JKY56) strains was examined by fluorescence microscopy. (B) Western blot analysis of GFP-Mth1 in wild-type and akr1Δ mutant strains. Yeast cells were grown as described in Figure 2.
Four different regions of Mth1 are required for glucose-induced degradation of Mth1
To identify the regions of Mth1 that are required for its glucose-induced degradation, we constructed a series of successive internal deletions of 10-50 residues covering the entire Mth1 sequence (Figure 6A). The amino acid positions between 20 and 80 of Mth1 were not deleted because this region is not well conserved in the Mth1 orthologs from other yeast species. Most of the deletion mutants of Mth1 protein were detectable by Western blotting, except two mutants that contain Δ235-262 (ID 11) and Δ298-319 (ID 13) (Figure 6B). Our results show that four internal deletions--Δ81-90 (ID 2), Δ118-138 (ID 4), 156-180 (ID 6), and Δ326-343 (ID 15)--render Mth1 resistant to degradation. These Mth1 mutant proteins are constitutively nuclear (data not shown) and significantly inhibit glucose induction of HXT1 expression (Figure 6C). Proteasomal degradation of proteins is a multi-step process, involving phosphorylation, ubiquitination, and degradation in the 26S proteasome. Thus, it is likely that these four regions are involved in at least one of these steps.
Identification of the functional regions of Mth1 required for glucose-dependent degradation. (A) Mth1 deletion mutants were generated to determine the regions responsible for degradation by successive 10-50 amino-acids deletion analysis. In total, 18 deletions were constructed. (B) The mutant Mth1 proteins containing internal deletions were expressed in the mth1Δ cells (YM6266) and detected by Western blotting using anti-GFP antibody. (C) Four deletion mutants of Mth1 inhibit glucose-induction of HXT1 expression. Four internal deletion constructs without the GFP moiety were coexpressed with the HXT1-lacZ reporter (pBM2636) in the absence (Gal, black bars) and presence (Glu, gray bars) of glucose.
Discussion
The yeast glucose sensors convert glucose binding events into an intracellular signal that leads to degradation of Mth1, which is known to require activity of the plasma membrane-tethered Yck1/2 [18]. Because both the glucose sensors and Yck1/2 are associated with the plasma membrane, it has been hypothesized that, upon glucose addition, Yck1/2 are activated through a direct interaction with the glucose sensors and phosphorylate Mth1, triggering its proteasomal degradation.
In this study, we provide several lines of evidence that Mth1 does not shuttle between the nucleus and cytoplasm and is degraded in the nucleus when glucose is present: 1) Mth1 is not excluded from the nucleus in response to glucose (Figure 2); 2) When engineered to be localized to the cytoplasm, Mth1 is degraded in the cytoplasm regardless of the presence of glucose and the glucose sensors (Figure 3); 3) Mth1 is not degraded when Grr1 is removed from the nucleus (Figure 4).
The majority of Yck1/2 targets are plasma membrane proteins. Yck1/2 are responsible for phosphorylating the PEST-like ubiquitination-endocytosis signal of the mating pheromone receptors [30], and the uracil permease Fur4 [31]. These kinases are also known to regulate the activity of the maltose permease Mal61 [32], the multidrug transporter Pdr5 [33], and plasma membrane H+-ATPase [34]. Another important target of Yck1/2 is Ptr3, a component of the yeast amino-acid signaling pathway. Extracellular amino acids trigger activation of the Ssy1-Ptr3-Ssy5 (SPS)-amino acid signaling pathway that leads to induction of endoproteolytic processing of Stp1 and Stp2, enabling them to enter the nucleus and induce expression of the amino acid permease genes [35–38]. This processing requires Yck1/2, the novel chymotrypsin like protease Ssy5, and SCFGrr1[36, 37]. Yck1/2 have been shown to phosphorylate the peripheral plasma membrane protein Ptr3 at Thr-525, increasing Ssy5C (C-terminal activity domain)-dependent proteolytic processing of Stp1 and Stp2 [39]. Mth1 is required to form a repression complex with Rgt1 on the HXT promoters [6] and appears not to contain endogenous NES-like motifs in its sequence (data not shown). These observations support the idea that Mth1 neither shuttles between the nucleus and cytoplasm nor is excluded from the nucleus. Therefore, it is unlikely that the plasma membrane-tethered Yck1/2 directly phosphorylate the nuclear-localized Mth1.
We also provide evidence that Mth1 degradation is not impaired by the mislocalization of Yck1/2 from the plasma membrane, suggesting that direct interaction between the glucose sensors and Yck1/2 is not required for Mth1 degradation (Figure 5). Although Yck1/2 have been reported to be necessary for Mth1 degradation [18], our results suggest that they do not exert their function through the glucose sensors. It is not known how the glucose binding to the glucose sensors is converted to an intracellular signal that leads to degradation of Mth1. In this regard, we surmise that there is a yet unidentified kinase that receives the glucose signal and converts it into an intracellular signal. In this scenario, the kinase is recruited to the glucose sensors upon glucose addition and phosphorylated by Yck1/2 at the plasma membrane. Finally, the kinase is translocated from the cytoplasm into the nucleus and catalyzes phosphorylation of Mth1, triggering its degradation by the 26S proteasome.
Conclusions
Glucose-induced, the glucose sensor-regulated Mth1 degradation occurs in the nucleus and requires neither nuclear export of Mth1 nor direct interaction between the glucose sensors and Yck1/2. The glucose sensors transmit their signal across the plasma membrane through a yet unidentified signaling component, not through Yck1/2.
Methods
Yeast strains and plasmids
Yeast strains were grown on YPD (1% yeast extract, 2% bacto-peptone, and 4% glucose) or synthetic yeast nitrogen base media (0.17% yeast nitrogen base with 0.5% ammonium sulfate) supplemented with the appropriate amino acids and carbon sources. The AKR1 gene was disrupted by homologous recombination using the NatMX cassette [40]. Plasmids that express the mutant Mth1 proteins with internal deletions were constructed by using the gap-repair technique [41]. To construct the GFP-MTH1 tagged with either the wild-type (NES, ELALKLAGLDIN) or mutated (NESm, ELALKLAGA DIN; L changed to A (Underlined)) leucine-rich nuclear export sequences of the yeast PKIα [42], synthetic oligonucleotides encoding the NES and NES(m) peptides were inserted between the GFP and MTH1 genes in the GFP-MTH1 construct [14]. Plasmids used in this study are listed in Table 1.
Fluorescence microscopy and FRAP (Fluorescence Recovery After Photobleaching)
GFP-fusion proteins expressed in yeast cells were visualized using a Zeiss LSM 510 META confocal laser scanning microscope with a 63× Plan-Apochromat 1.4 NA Oil DIC objective lens (Zeiss) [17]. All images documenting GFP localization were acquired with the Zeiss LSM 510 software version 3.2. For FRAP of GFP-Mth1, one of the foci was bleached with a laser pulse and the subsequent recovery of fluorescence was monitored [17].
Western blotting
Western blotting was performed as described previously [6]. Briefly, 5 ml of yeast cells (O.D600 = 1.2) were collected by centrifugation at 3,000 rpm in a table-top centrifuge for 5 min. The cell pellets were resuspended in 100 μl of SDS-buffer (50 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, 5% β-mercaptoethanol) and boiled for 5 min. After the lysates were cleared by centrifugation at 12,000 rpm for 10 min., soluble proteins were resolved by SDS-PAGE and transferred to PVDF membrane (Millipore). The membranes were incubated with appropriate antibodies in TBST buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.05% Tween-20) and proteins were detected by the enhanced chemiluminescence (ECL) system (Pierce).
β-galactosidase assay
To assay β-galactosidase activity with yeast cells expressing appropriate lacZ reporters, yeast cells were grown to mid-log phase and assay was performed as described previously [6]. Results were reported in Miller Units [(1,000 × OD420)/(T × V × OD600), where OD420 was the optical density at 420 nm, T was the incubation time in minutes, and V is the volume of cells in milliliters]. The reported enzyme activities were averages of results from triplicates of three different transformants.
References
Ozcan S, Johnston M: Function and regulation of yeast hexose transporters. Microbiol Mol Biol Rev. 1999, 63 (3): 554-569.
Rolland F, Winderickx J, Thevelein JM: Glucose-sensing and -signalling mechanisms in yeast. FEMS Yeast Res. 2002, 2 (2): 183-201.
Johnston M, Kim JH: Glucose as a hormone: receptor-mediated glucose sensing in the yeast Saccharomyces cerevisiae. Biochem Soc Trans. 2005, 33 (Pt 1): 247-252.
Ozcan S, Johnston M: Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol Cell Biol. 1995, 15 (3): 1564-1572.
Ozcan S, Johnston M: Two different repressors collaborate to restrict expression of the yeast glucose transporter genes HXT2 and HXT4 to low levels of glucose. Mol Cell Biol. 1996, 16 (10): 5536-5545.
Kim JH, Polish J, Johnston M: Specificity and regulation of DNA binding by the yeast glucose transporter gene repressor Rgt1. Mol Cell Biol. 2003, 23 (15): 5208-5216. 10.1128/MCB.23.15.5208-5216.2003.
Kim JH: Immobilized DNA-binding assay, an approach for in vitro DNA-binding assay. Anal Biochem. 2004, 334 (2): 401-402. 10.1016/j.ab.2004.06.045.
Polish JA, Kim JH, Johnston M: How the Rgt1 transcription factor of Saccharomyces cerevisiae is regulated by glucose. Genetics. 2005, 169 (2): 583-594. 10.1534/genetics.104.034512.
Tomas-Cobos L, Sanz P: Active Snf1 protein kinase inhibits expression of the Saccharomyces cerevisiae HXT1 glucose transporter gene. Biochem J. 2002, 368 (Pt 2): 657-663. 10.1042/BJ20020984.
Lakshmanan J, Mosley AL, Ozcan S: Repression of transcription by Rgt1 in the absence of glucose requires Std1 and Mth1. Curr Genet. 2003, 44 (1): 19-25. 10.1007/s00294-003-0423-2.
Mosley AL, Lakshmanan J, Aryal BK, Ozcan S: Glucose-mediated phosphorylation converts the transcription factor Rgt1 from a repressor to an activator. J Biol Chem. 2003, 278 (12): 10322-10327. 10.1074/jbc.M212802200.
Flick KM, Spielewoy N, Kalashnikova TI, Guaderrama M, Zhu Q, Chang HC, Wittenberg C: Grr1-dependent inactivation of Mth1 mediates glucose-induced dissociation of Rgt1 from HXT gene promoters. Mol Biol Cell. 2003, 14 (8): 3230-3241. 10.1091/mbc.E03-03-0135.
Spielewoy N, Flick K, Kalashnikova TI, Walker JR, Wittenberg C: Regulation and recognition of SCFGrr1 targets in the glucose and amino acid signaling pathways. Mol Cell Biol. 2004, 24 (20): 8994-9005. 10.1128/MCB.24.20.8994-9005.2004.
Kim JH, Brachet V, Moriya H, Johnston M: Integration of transcriptional and posttranslational regulation in a glucose signal transduction pathway in Saccharomyces cerevisiae. Eukaryot Cell. 2006, 5 (1): 167-173. 10.1128/EC.5.1.167-173.2006.
Ozcan S, Dover J, Rosenwald AG, Wolfl S, Johnston M: Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc Natl Acad Sci USA. 1996, 93 (22): 12428-12432. 10.1073/pnas.93.22.12428.
Ozcan S, Dover J, Johnston M: Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. Embo J. 1998, 17 (9): 2566-2573. 10.1093/emboj/17.9.2566.
Pasula S, Jouandot D, Kim JH: Biochemical evidence for glucose-independent induction of HXT expression in Saccharomyces cerevisiae. FEBS Lett. 2007, 581 (17): 3230-3234. 10.1016/j.febslet.2007.06.013.
Moriya H, Johnston M: Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc Natl Acad Sci USA. 2004, 101 (6): 1572-1577. 10.1073/pnas.0305901101.
Schmidt MC, McCartney RR, Zhang X, Tillman TS, Solimeo H, Wolfl S, Almonte C, Watkins SC: Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1999, 19 (7): 4561-4571.
Schulte F, Wieczorke R, Hollenberg CP, Boles E: The HTR1 gene is a dominant negative mutant allele of MTH1 and blocks Snf3- and Rgt2-dependent glucose signaling in yeast. J Bacteriol. 2000, 182 (2): 540-542. 10.1128/JB.182.2.540-542.2000.
Lafuente MJ, Gancedo C, Jauniaux JC, Gancedo JM: Mth1 receives the signal given by the glucose sensors Snf3 and Rgt2 in Saccharomyces cerevisiae. Mol Microbiol. 2000, 35 (1): 161-172. 10.1046/j.1365-2958.2000.01688.x.
Kaniak A, Xue Z, Macool D, Kim JH, Johnston M: Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae. Eukaryot Cell. 2004, 3 (1): 221-231. 10.1128/EC.3.1.221-231.2004.
Ozcan S, Freidel K, Leuker A, Ciriacy M: Glucose uptake and catabolite repression in dominant HTR1 mutants of Saccharomyces cerevisiae. J Bacteriol. 1993, 175 (17): 5520-5528.
Stochaj U, Osborne M, Kurihara T, Silver P: A yeast protein that binds nuclear localization signals: purification localization, and antibody inhibition of binding activity. J Cell Biol. 1991, 113 (6): 1243-1254. 10.1083/jcb.113.6.1243.
Blondel M, Bach S, Bamps S, Dobbelaere J, Wiget P, Longaretti C, Barral Y, Meijer L, Peter M: Degradation of Hof1 by SCF(Grr1) is important for actomyosin contraction during cytokinesis in yeast. Embo J. 2005, 24 (7): 1440-1452. 10.1038/sj.emboj.7600627.
Li FN, Johnston M: Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: coupling glucose sensing to gene expression and the cell cycle. Embo J. 1997, 16 (18): 5629-5638. 10.1093/emboj/16.18.5629.
Feng Y, Davis NG: Akr1p and the type I casein kinases act prior to the ubiquitination step of yeast endocytosis: Akr1p is required for kinase localization to the plasma membrane. Mol Cell Biol. 2000, 20 (14): 5350-5359. 10.1128/MCB.20.14.5350-5359.2000.
Roth AF, Feng Y, Chen L, Davis NG: The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J Cell Biol. 2002, 159 (1): 23-28. 10.1083/jcb.200206120.
Babu P, Deschenes RJ, Robinson LC: Akr1p-dependent palmitoylation of Yck2p yeast casein kinase 1 is necessary and sufficient for plasma membrane targeting. J Biol Chem. 2004, 279 (26): 27138-27147. 10.1074/jbc.M403071200.
Hicke L, Zanolari B, Riezman H: Cytoplasmic tail phosphorylation of the alpha-factor receptor is required for its ubiquitination and internalization. J Cell Biol. 1998, 141 (2): 349-358. 10.1083/jcb.141.2.349.
Marchal C, Haguenauer-Tsapis R, Urban-Grimal D: A PEST-like sequence mediates phosphorylation and efficient ubiquitination of yeast uracil permease. Mol Cell Biol. 1998, 18 (1): 314-321.
Gadura N, Robinson LC, Michels CA: Glc7-Reg1 phosphatase signals to Yck1,2 casein kinase 1 to regulate transport activity and glucose-induced inactivation of Saccharomyces maltose permease. Genetics. 2006, 172 (3): 1427-1439. 10.1534/genetics.105.051698.
Decottignies A, Owsianik G, Ghislain M: Casein kinase I-dependent phosphorylation and stability of the yeast multidrug transporter Pdr5p. J Biol Chem. 1999, 274 (52): 37139-37146. 10.1074/jbc.274.52.37139.
Estrada E, Agostinis P, Vandenheede JR, Goris J, Merlevede W, Francois J, Goffeau A, Ghislain M: Phosphorylation of yeast plasma membrane H+-ATPase by casein kinase I. J Biol Chem. 1996, 271 (50): 32064-32072. 10.1074/jbc.271.50.32064.
Klasson H, Fink GR, Ljungdahl PO: Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol Cell Biol. 1999, 19 (8): 5405-5416.
Andreasson C, Ljungdahl PO: Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Genes Dev. 2002, 16 (24): 3158-3172. 10.1101/gad.239202.
Abdel-Sater F, El Bakkoury M, Urrestarazu A, Vissers S, Andre B: Amino acid signaling in yeast: casein kinase I and the Ssy5 endoprotease are key determinants of endoproteolytic activation of the membrane-bound Stp1 transcription factor. Mol Cell Biol. 2004, 24 (22): 9771-9785. 10.1128/MCB.24.22.9771-9785.2004.
Gaber RF, Ottow K, Andersen HA, Kielland-Brandt MC: Constitutive and hyperresponsive signaling by mutant forms of Saccharomyces cerevisiae amino acid sensor Ssy1. Eukaryot Cell. 2003, 2 (5): 922-929. 10.1128/EC.2.5.922-929.2003.
Liu Z, Thornton J, Spirek M, Butow RA: Activation of the SPS amino acid-sensing pathway in Saccharomyces cerevisiae correlates with the phosphorylation state of a sensor component, Ptr3. Mol Cell Biol. 2008, 28 (2): 551-563. 10.1128/MCB.00929-07.
Goldstein AL, Pan X, McCusker JH: Heterologous URA3MX cassettes for gene replacement in Saccharomyces cerevisiae. Yeast. 1999, 15 (6): 507-511. 10.1002/(SICI)1097-0061(199904)15:6<507::AID-YEA369>3.0.CO;2-P.
Ma H, Kunes S, Schatz PJ, Botstein D: Plasmid construction by homologous recombination in yeast. Gene. 1987, 58 (2-3): 201-216. 10.1016/0378-1119(87)90376-3.
Feng Y, Davis NG: Akr1p and the type I casein kinases act prior to the ubiquitination step of yeast endocytosis: Akr1p is required for kinase localization to the plasma membrane. Mol Cell Biol. 2000, 20 (14): 5350-5359. 10.1128/MCB.20.14.5350-5359.2000.
Acknowledgements
This work was supported by R01GM087470 (JHK) from the National Institute of General Medical Sciences (NIGMS) and RR016476-08 (MS-INBRE) from the National Center for Research Resources (NCRR).
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
SP and SC were responsible for all the experimental work. JC conceived of the study and was involved in writing the manuscript. JK participated in study design and analyses and wrote the manuscript with input from SP. All authors have read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Pasula, S., Chakraborty, S., Choi, J.H. et al. Role of casein kinase 1 in the glucose sensor-mediated signaling pathway in yeast. BMC Cell Biol 11, 17 (2010). https://doi.org/10.1186/1471-2121-11-17
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2121-11-17