Abstract
Background
Segmentation of computed tomography (CT) images provides quantitative data on body tissue composition, which may greatly impact the development and progression of diseases such as type 2 diabetes mellitus and cancer. We aimed to evaluate the inter- and intraobserver variation of semiautomated segmentation, to assess whether multiple observers may interchangeably perform this task.
Methods
Anonymised, unenhanced, single mid-abdominal CT images were acquired from 132 subjects from two previous studies. Semiautomated segmentation was performed using a proprietary software package. Abdominal muscle compartment (AMC), inter- and intramuscular adipose tissue (IMAT), visceral adipose tissue (VAT) and subcutaneous adipose tissue (SAT) were identified according to pre-established attenuation ranges. The segmentation was performed by four observers: an oncology resident with extensive training and three radiographers with a 2-week training programme. To assess interobserver variation, segmentation of each CT image was performed individually by two or more observers. To assess intraobserver variation, three of the observers did repeated segmentations of the images. The distribution of variation between subjects, observers and random noise was estimated by a mixed effects model. Inter- and intraobserver correlation was assessed by intraclass correlation coefficient (ICC).
Results
For all four tissue compartments, the observer variations were far lower than random noise by factors ranging from 1.6 to 3.6 and those between subjects by factors ranging from 7.3 to 186.1. All interobserver ICC was ≥ 0.938, and all intraobserver ICC was ≥ 0.996.
Conclusions
Body composition segmentation showed a very low level of operator dependability. Multiple observers may interchangeably perform this task with highly reproducible results.
Similar content being viewed by others
Key points
-
Body composition data may predict development and progression and aid treatment of noncommunicable diseases such as type 2 diabetes mellitus and cancer.
-
Semiautomated body composition segmentation on abdominal CT images showed a very low inter- and intraobserver variation (intraclass correlation coefficient from 0.938 to 0.996).
-
Semiautomated body composition segmentation may be performed by non-radiologists after a short period of training.
Background
Computed tomography (CT) is part of the routine work-up in many patient groups. With special image segmentation software, high-precision data on body composition, i.e. the quantification and distribution of different tissues, may be extracted from these images [1].
Body composition states such as obesity and sarcopenia are associated with the risk of development and progression of noncommunicable diseases as well as overall survival [2,3,4,5,6]. Excess adipose tissue in the abdominal region increases the risk of type 2 diabetes mellitus (T2DM), cardiometabolic diseases and some cancers [2, 7]. Sarcopenia is a recognised diabetic and oncologic complication, and insulin resistance is a central mechanism both in sarcopenia and obesity-related diseases [8,9,10,11,12]. Central obesity with sarcopenia, i.e. sarcopenic obesity, may increase the effects on metabolic disorders, cardiovascular diseases and mortality [13].
Image segmentation is increasingly used as a research tool in areas such as oncology, endocrinology, cardiovascular disease, nutrition, obesity and ageing [1, 14, 15]. Semiautomated methods are faster, easier and more versatile than manual delineation and without loss of precision [14, 16,17,18]. Segmentation is validated against cadaver studies and offers advantages to dual-energy X-ray absorptiometry scans and bioelectric impedance analysis [19,20,21].
Since the cross-sectional areas at the third lumbar vertebra are linearly related to whole body mass of muscle, visceral adipose tissue (VAT) and subcutaneous adipose tissue (SAT), a single axial image is often acquired in research settings to reduce cost or radiation exposure [1, 22, 23].
Body composition analysis is also valuable in current clinical practice such as identification of cachexia in patients with cancer and is included in the newly published GLIM (Global Leadership Initiative on Malnutrition) criteria for malnutrition [24,25,26,27,28]. Assessment of nutritional status in patients with noncommunicable diseases such as T2DM and cancer may facilitate personalised and precision medicine and greatly impact treatment and prognosis [2, 7, 13, 25].
In order to acquire large amounts of data, for clinical or research purposes, the process of image segmentation and analysis must be quick and precise. It would be a practical advantage if image segmentation could be performed by a group of personnel interchangeably rather than one dedicated person.
Our aim was to evaluate the inter- and intraobserver variation of semiautomated body composition segmentation of CT images in both healthy and diabetic subjects, to assess whether multiple observers may interchangeably perform segmentation with comparable results.
Methods
Ethics and study population
Participant consent and approval from the Regional Committee for Medical and Health Research Ethics were previously obtained.
We obtained CT images of 41 subjects with T2DM enrolled in the Diabetes-study [29] and 91 healthy male subjects from the INFO-study [30]. The Diabetes-study population was aged 29 to 45 years (median 41), 49% males, with a mean body mass index (BMI) of 34.0 kg/m2. The INFO-study population was aged 38 to 45 years (median 40), all males, with a mean BMI of 26.4 kg/m2.
Computed tomography
Anonymised, unenhanced single abdominal CT images were acquired. In the Diabetes-study, CT images were obtained with a Somatom Volume Zoom, 4-slice CT scanner (Siemens Healthineers, Erlangen, Germany) at 5 cm above L4/L5 level in women and 10 cm above L4/L5 level in men with 120 kVp, 100 mAs and slice thickness 4 mm. In the INFO-study, CT images were obtained with a Somatom Sensation 64, 64-slice scanner (Siemens Healthineers, Erlangen, Germany) at L3/L4 level with 120 kVp, 200 mAs and slice thickness 5 mm.
Image analysis
Semiautomated body composition segmentation of the CT images was performed with the SliceOmatic software package (v 5.0 rev 7b, Tomovision, Magog, QC, Canada).
Body composition segmentation included four tissue compartments: abdominal muscle compartment (AMC), inter- and intramuscular adipose tissue (IMAT), VAT and SAT.
The segmentation was performed by four observers: three radiographers (radiology technicians) and one oncology resident. The resident had previously received training in the use of SliceOmatic at the University of Alberta hospital, Edmonton, AB, Canada. Over the course of 2 weeks, the resident held three 1-h teaching sessions for the technicians. This was followed by 7 to 12 h of practical training in the use of the software with individual feedback from the resident and two radiologists supervising the study.
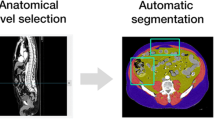
Segmentation was performed according to the Alberta protocol, defined and used at the Alberta Hospital (AB, Canada), as shown in Fig. 1 [31]. By this definition, AMC is muscle tissue free of adipose tissue, not anatomical muscle which may include intramuscular fat, and IMAT was segmented separately as adipose tissue within the muscle fasciae. For each tissue, segmentation was restricted to the following predefined attenuation ranges: − 29 to 150 Hounsfield Units (HU) for AMC, − 190 to − 30 HU for IMAT and SAT and − 150 to − 50 HU for VAT [19, 31, 32].
The three radiographers (observers 1, 2 and 3) performed segmentation of all the CT images from both studies. They were organised into alternating pairs so that each image was analysed independently by two radiographers. To evaluate intraobserver variation, the three radiographers performed a second segmentation of the same images from the Diabetes-study, after a 1-month delay. The oncology resident (observer 4) performed segmentation of all images from the Diabetes-study. The observers were blinded to each other’s results and their own previous results. A flow chart describing the distribution of performed segmentations between the observers is shown in Fig. 2.
From the different modes, available in the SliceOmatic software, the observers utilised the Region growing-mode with the Paint and Grow 2D options. This mode allowed the users to delineate the different types of tissue based on predefined attenuation ranges. The tissue compartments were tagged in a specific order, starting with AMC followed by SAT, VAT and IMAT. Although the software could delineate these four compartments semiautomatically using the Grow 2D option, all the segmented compartments were manually adjusted in each image with the Paint tool to ensure that the compartments had been segmented correctly, especially around the muscle with nearby tissues of similar density such as bowels or kidneys, but also around vertebra and IMAT.
Exclusion criteria
CT images with inferior quality due to noise, respiratory artefacts or other movement artefacts were excluded from analysis. Images where AMC was cut from the field of view (FOV) bilaterally were also excluded. In images where the oblique or transverse abdominal muscles were cut unilaterally from the FOV, AMC was estimated by segmentation of the contralateral AMC multiplied by two. In images where SAT or VAT was cut unilaterally or bilaterally from the FOV, segmentation of the affected tissue compartment was not performed.
Statistical analysis
Segmentation data in square centimetres per tissue compartment for each image was exported from SliceOmatic to Microsoft Excel (version 14.0, Microsoft Corporation, Redmond, WA, USA) and analysed in IBM SPSS Statistics (version 23, IBM Corporation, Armonk, NY, USA) and R (version 3.3, www.r-project.org).
Descriptive statistics are presented as median, minimum, maximum and percentiles. Normal distribution of measurement data was evaluated with Q-Q plots and Shapiro-Wilk tests.
For each of the four tissue compartments, the underlying variations in the measurement data between individual subjects, individual observers and residual variation (random noise) were analysed with mixed effects models. Different levels of variation and interactions between variations were evaluated in three different potential mixed effects models named A, B and C. All models included variation between individual subjects and random noise. Additionally, model A included general observer to observer variation, model B included systematic variation between observers in how segmentation was performed and model C included both sources of variation.
Parameters for each model were estimated by restricted maximum likelihood. Model fit for the three models was evaluated by Akaike information criterion (AIC), and results from the best fitting model presented as estimated standard deviations with 95% confidence intervals.
Mixed effects models assume normally distributed residuals, which were not present in the measurement data as seen in the Q-Q plots. The efficiency of the applied estimation routine was evaluated by simulations, and a robustness analysis was performed on data transformed to achieve normally distributed residuals.
In addition to the mixed effects model estimates, intraclass correlation coefficients (ICCs) were calculated. Two-way random effect ICC was used for overall variation in segmentation results, and single measurement ICC for intraobserver variation. Confidence intervals for the two-way ICC were based on a random effects model with percentile bootstrap confidence intervals based on 10,000 replications randomly sampling subjects and observers.
Subgroup analyses were performed for the Diabetes-study and the INFO-study in order to explore differences in variations between the subject groups.
Results
Body composition segmentation was performed on 120 of 132 CT images. Eight were excluded from analysis due to compartments being cut from the field of view, two due to respiratory or movement artefacts and two due to image noise. From the 120 images, we acquired 346 sets of segmentation data for AMC, IMAT and VAT and 338 sets for SAT.
Q-Q plots showed that none of the segmentation data were normally distributed (all Shapiro-Wilk tests p < 0.010). Descriptive statistics of segmentation data of the four tissue compartments are shown in Table 1.
For each of the four tissue compartments, AIC showed best fit for model A, modelling only general observer to observer variation (Table 2). Therefore, systematic variation between observers in how segmentation was performed was not included in the final mixed effects model.
For both studies combined, the variations between observers were consistently less than variation between subjects for all four tissue compartments by a factor of 7.3 to 186.1. Variations between observers were also less than random noise by a factor of 1.6 to 3.6 (Table 3, Fig. 3). Mixed effects model analysis yielded results with very similar interpretations on non-transformed data (Table 3) and data transformed to achieve normally distributed residuals (Additional file 1: Table S1). Simulations of non-normal distributed data showed efficient restricted maximum likelihood estimates also with similar violations of the normally distribution assumption as seen in our data (results not presented).
For both studies combined, the interobserver ICC ranged from 0.938 to 1.000 for all four compartments, with IMAT scoring the lowest (Table 4). All intraobserver ICC ranged from 0.996 to 1.000.
Subgroup analysis
For the Diabetes-study, the variations between observers were consistently less than the variations between subjects for all four tissue compartments by a factor of 10.0 to 212.1 (Table 3). The variations between observers were also less than random noise by a factor of 1.8 to 4.0. For all four compartments, the interobserver ICC ranged from 0.961 to 1.00 (Table 4).
For the INFO-study, the variations between observers were consistently less than the variations between subjects for all four tissue compartments by a factor of 3.1 to 99.7. The variations between observers were also less than random noise by a factor of 1.2 to 3.7. For IMAT, the interobserver ICC was 0.759, and for the remaining three compartments ICC ranged from 0.987 to 1.00.
Discussion
Our results show that after a short period of training non-radiologist physicians and radiographers can perform semiautomated segmentation of body composition on abdominal CT images with close to identical results.
Van Vugt et al. [17] showed similar results with close to perfect ICCs for inter- and intraobserver agreement. However, their observers had extensive experience in skeletal muscle and adipose tissue area measurement, whereas our inexperienced technicians produced similar results with only a short period of training.
Our results underline that the intuitive nature of the software and the standardised process of semiautomated segmentation produces consistent results even when performed by operators with less radiological experience. This facilitates segmentation of larger number of images without loss of data quality, which increases the value of this method, whether used for patient follow-up in clinical settings or measurements of research endpoints.
From the three mixed effects models, AIC consistently showed superior fit for model A, with only general observer variation. This strengthens the assumption that there was a variation between observers, but no systematic variation between observers in how segmentation was performed.
Both the mixed effects model and the ICC showed, consistently for all four tissue compartments, that observer variation was less than variation due to random noise and negligible compared to the variation between individual subjects. Consequently, a single dedicated person is not necessary for the acquisition of reliable segmentation data, and segmentation may be performed interchangeably by any member of a group of trained radiographers. This allows for greater flexibility and makes feasible the acquisition of greater amounts of data or even the future adoption into clinical practice.
In our experience, segmentation of AMC and SAT was relatively straightforward, which is supported by our data. The slightly higher observer variation observed for VAT in the mixed effects model may indicate that segmentation of this compartment is more demanding than that of AMC and SAT, which is in line with our experience. Structures in the same anatomical space as VAT, such as the viscera, the mesentery, the intestinal wall and fatty contents of the intestines, may complicate segmentation.
The slightly lower interobserver ICC for IMAT may primarily be due to the relatively small variation in this tissue compartment between subjects compared to AMC, VAT and SAT. Contributing factors may be the relatively small area of IMAT in each image and a less stringent definition of this compartment. These explanations are further supported by the subgroup analyses specifically showing a lower interobserver ICC for IMAT in the INFO-study subject group, although with a wide confidence interval. The median area of IMAT in the INFO-study was approx. one third compared to the Diabetes-study. Hence, the relative effect of observer variation was larger in the IMAT measurement resulting in a lower ICC. However, in our opinion, the subgroup analyses confirm that our results are valid for both groups separately.
We decided to present the results of the mixed effects model even though the requirement of normally distributed residuals was not met. Maximum likelihood estimation on medium-sized datasets tends to give reasonable estimates even with some violations of the modelling assumptions, which we confirmed with simulations. The analysis of non-transformed data allowed us to present the results as standard deviations, which could be compared across the segmented compartments and are easier to understand and interpret.
In order to control the robustness of the method, we carried out a sensitivity analysis with the same model on transformed data with approximately normally distributed residuals. This sensitivity analysis showed similar results, confirming the validity of our results, though in a format which is more difficult to understand and interpret.
Limitations of our study should be taken into account. Our study was limited to one, specific software, and our results may not apply perfectly to other segmentation tools. Furthermore, we did not account for variation associated with image acquisition such as the level or angle of the CT slice [1]. In addition, the exclusion of 12 images due to artefacts and noise show that not all acquired images are suitable for segmentation and that standardised acquisition protocols are necessary. Furthermore, the time spent performing segmentation was not specifically measured.
We conclude that semiautomated body composition segmentation using SliceOmatic showed a very low level of operator dependability. Hence, multiple observers may interchangeably perform body composition segmentation of abdominal CT with close to identical results in a clinical or research setting.
Availability of data and materials
The datasets used and analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AIC:
-
Akaike information criterion
- AMC:
-
Abdominal muscle compartment
- BMI:
-
Body mass index
- CT:
-
Computed tomography
- ICC:
-
Intraclass correlation coefficient
- IMAT:
-
Inter- and intramuscular adipose tissue
- SAT:
-
Subcutaneous adipose tissue
- T2DM:
-
Type 2 diabetes mellitus
- VAT:
-
Visceral adipose tissue
References
MacDonald AJ, Greig CA, Baracos V (2011) The advantages and limitations of cross-sectional body composition analysis. Curr Opin Support Palliat Care 5:342–349. https://doi.org/10.1097/SPC.0b013e32834c49eb
Nishida C, Ko GT, Kumanyika S (2010) Body fat distribution and noncommunicable diseases in populations: overview of the 2008 WHO Expert Consultation on Waist Circumference and Waist-Hip Ratio. Eur J Clin Nutr 64:2–5. https://doi.org/10.1038/ejcn.2009.139
Prado CM, Lieffers JR, McCargar LJ et al (2008) Prevalence and clinical implications of sarcopenic obesity in patients with solid tumours of the respiratory and gastrointestinal tracts: a population-based study. Lancet Oncol 9:629–635. https://doi.org/10.1016/S1470-2045(08)70153-0
van Kruijsdijk RC, van der Wall E, Visseren FL (2009) Obesity and cancer: the role of dysfunctional adipose tissue. Cancer Epidemiol Biomarkers Prev 18:2569–2578. https://doi.org/10.1158/1055-9965.EPI-09-0372
World Cancer Research Fund International/American Institute for Cancer Research (2017) Continuous update project report: diet, nutrition, physical activity and colorectal cancer. wcrf.org/colorectal-cancer-2017. Accessed June:2019
Cruz-Jentoft AJ, Bahat G, Bauer J et al (2019) Sarcopenia: revised European consensus on definition and diagnosis. Age Ageing 48:16–31. https://doi.org/10.1093/ageing/afy169
Martin FP, Montoliu I, Collino S et al (2013) Topographical body fat distribution links to amino acid and lipid metabolism in healthy obese women [corrected]. PLoS One 8:e73445. https://doi.org/10.1371/journal.pone.0073445
Bitar MS, Nader J, Al-Ali W, Al Madhoun A, Arefanian H, Al-Mulla F (2018) Hydrogen sulfide donor NaHS improves metabolism and reduces muscle atrophy in type 2 diabetes: implication for understanding sarcopenic pathophysiology. Oxid Med Cell Longev 2018:6825452. https://doi.org/10.1155/2018/6825452
Tanaka KI, Kanazawa I, Notsu M, Sugimoto T (2018) Higher serum uric acid is a risk factor of reduced muscle mass in men with type 2 diabetes mellitus. Exp Clin Endocrinol Diabetes. https://doi.org/10.1055/a-0805-2197
Umegaki H (2015) Sarcopenia and diabetes: hyperglycemia is a risk factor for age-associated muscle mass and functional reduction. J Diabetes Investig 6:623–624. https://doi.org/10.1111/jdi.12365
Hilmi M, Jouinot A, Burns R et al (2019) Body composition and sarcopenia: the next-generation of personalized oncology and pharmacology? Pharmacol Ther 196:135–159. https://doi.org/10.1016/j.pharmthera.2018.12.003
Marty E, Liu Y, Samuel A, Or O, Lane J (2017) A review of sarcopenia: enhancing awareness of an increasingly prevalent disease. Bone 105:276–286. https://doi.org/10.1016/j.bone.2017.09.008
Kreidieh D, Itani L, El Masri D, Tannir H, Citarella R, El Ghoch M (2018) Association between sarcopenic obesity, type 2 diabetes, and hypertension in overweight and obese treatment-seeking adult women. J Cardiovasc Dev Dis 5:51. https://doi.org/10.3390/jcdd5040051
Latini F, Larsson EM, Ryttlefors M (2017) Rapid and accurate MRI segmentation of peritumoral brain edema in meningiomas. Clin Neuroradiol 27:145–152. https://doi.org/10.1007/s00062-015-0481-0
Shahedi M, Halicek M, Guo R, Zhang G, Schuster DM, Fei B (2018) A semiautomatic segmentation method for prostate in CT images using local texture classification and statistical shape modeling. Med Phys 45:2527–2541. https://doi.org/10.1002/mp.12898
Newman D, Kelly-Morland C, Leinhard OD et al (2016) Test-retest reliability of rapid whole body and compartmental fat volume quantification on a widebore 3T MR system in normal-weight, overweight, and obese subjects. J Magn Reson Imaging 44:1464–1473. https://doi.org/10.1002/jmri.25326
van Vugt JL, Levolger S, Gharbharan A et al (2017) A comparative study of software programmes for cross-sectional skeletal muscle and adipose tissue measurements on abdominal computed tomography scans of rectal cancer patients. J Cachexia Sarcopenia Muscle 8:285–297. https://doi.org/10.1002/jcsm.12158
Ozola-Zalite I, Mark EB, Gudauskas T et al (2019) Reliability and validity of the new VikingSlice software for computed tomography body composition analysis. Eur J Clin Nutr 73:54–61. https://doi.org/10.1038/s41430-018-0110-5
Mitsiopoulos N, Baumgartner RN, Heymsfield SB, Lyons W, Gallagher D, Ross R (1998) Cadaver validation of skeletal muscle measurement by magnetic resonance imaging and computerized tomography. J Appl Physiol (1985) 85:115–122. https://doi.org/10.1152/jappl.1998.85.1.115
Nelson ME, Fiatarone MA, Layne JE et al (1996) Analysis of body-composition techniques and models for detecting change in soft tissue with strength training. Am J Clin Nutr 63:678–686. https://doi.org/10.1093/ajcn/63.5.678
Bredella MA, Ghomi RH, Thomas BJ et al (2010) Comparison of DXA and CT in the assessment of body composition in premenopausal women with obesity and anorexia nervosa. Obesity (Silver Spring) 18:2227–2233. https://doi.org/10.1038/oby.2010.5
Shen W, Punyanitya M, Wang Z et al (2004) Total body skeletal muscle and adipose tissue volumes: estimation from a single abdominal cross-sectional image. J Appl Physiol (1985) 97:2333–2338. https://doi.org/10.1152/japplphysiol.00744.2004
Lee S, Janssen I, Ross R (2004) Interindividual variation in abdominal subcutaneous and visceral adipose tissue: influence of measurement site. J Appl Physiol (1985) 97:948–954. https://doi.org/10.1152/japplphysiol.01200.2003
Agustsson T, Wikrantz P, Ryden M, Brismar T, Isaksson B (2012) Adipose tissue volume is decreased in recently diagnosed cancer patients with cachexia. Nutrition 28:851–855. https://doi.org/10.1016/j.nut.2011.11.026
Trestini I, Carbognin L, Monteverdi S et al (2018) Clinical implication of changes in body composition and weight in patients with early-stage and metastatic breast cancer. Crit Rev Oncol Hematol 129:54–66. https://doi.org/10.1016/j.critrevonc.2018.06.011
Bowden JCS, Williams LJ, Simms A et al (2017) Prediction of 90 day and overall survival after chemoradiotherapy for lung cancer: role of performance status and body composition. Clin Oncol (R Coll Radiol) 29:576–584. https://doi.org/10.1016/j.clon.2017.06.005
Jensen GL, Cederholm T, Correia M et al (2019) GLIM criteria for the diagnosis of malnutrition: a consensus report from the global clinical nutrition community. JPEN J Parenter Enteral Nutr 43:32–40. https://doi.org/10.1002/jpen.1440
Kazemi-Bajestani SM, Mazurak VC, Baracos V (2016) Computed tomography-defined muscle and fat wasting are associated with cancer clinical outcomes. Semin Cell Dev Biol 54:2–10. https://doi.org/10.1016/j.semcdb.2015.09.001
Wium C, Eggesbo HB, Ueland T et al (2014) Adipose tissue distribution in relation to insulin sensitivity and inflammation in Pakistani and Norwegian subjects with type 2 diabetes. Scand J Clin Lab Invest 74:700–707. https://doi.org/10.3109/00365513.2014.953571
Skarn SN, Eggesbo HB, Flaa A et al (2016) Predictors of abdominal adipose tissue compartments: 18-year follow-up of young men with and without family history of diabetes. Eur J Intern Med 29:26–31. https://doi.org/10.1016/j.ejim.2015.11.027
Prado CM, Baracos VE, McCargar LJ et al (2007) Body composition as an independent determinant of 5-fluorouracil-based chemotherapy toxicity. Clin Cancer Res 13:3264–3268. https://doi.org/10.1158/1078-0432.CCR-06-3067
Miller KD, Jones E, Yanovski JA, Shankar R, Feuerstein I, Falloon J (1998) Visceral abdominal-fat accumulation associated with use of indinavir. Lancet 351:871–875. https://doi.org/10.1016/S0140-6736(97)11518-5
Acknowledgements
Cecilie Wium, Sigrid Skårn Nordang and Tonje Amb Aksnes, investigators of the Diabetes-study and INFO-study.
Funding
The study received no external funding and was carried out with the resources of the author affiliated institutions.
Author information
Authors and Affiliations
Contributions
LJK, MH, HBE, HWF and PML contributed to the design of the study. AMF, LKK, KP and MH performed the body composition segmentation. LJK, MH, GS, HBE, HWF, HBH and PML contributed to the data analysis and interpretation of the results. LJK drafted the manuscript. All authors revised the manuscript and approved the final version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Participant consent and approval from the Norwegian South-East Regional Committee for Medical and Health Research Ethics (2009/157 and 2010/3339) were previously obtained.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Table S1. Distribution of variation between subjects, observers, and random noise for transformed, normally distributed data. (DOCX 18 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kjønigsen, L.J., Harneshaug, M., Fløtten, AM. et al. Reproducibility of semiautomated body composition segmentation of abdominal computed tomography: a multiobserver study. Eur Radiol Exp 3, 42 (2019). https://doi.org/10.1186/s41747-019-0122-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41747-019-0122-5