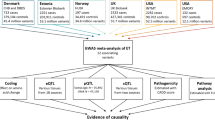
Abstract
Background
The aim of the study was to investigate the genetic risk factors of essential tremor (ET) in Chinese Population.
Methods
A total of 225 ET patients (25 ET patients also had restless legs syndrome (RLS) and were excluded from final analysis) and 229 controls were recruited. The diagnosis of ET was based on the Consensus Statement of the Movement Disorders Society on tremor. Polymerase chain reaction (PCR) and sequencing were used to detect 12 single nucleotide polymorphisms (SNPs) in seven candidate genes for RLS (HMOX1, HMOX2, VDR, IL17A, IL1B, NOS1 and ADH1B).
Results
We found that one SNP was associated with the risk of ET in Chinese population after adjusting for age and gender: rs1143633 of IL1B (odds ratio [OR] =2.57, p = 0.003, recessive model), and the statistical result remained significant after Bonferroni correction. Then, we performed a query in Genotype-tissue Expression (GTEx), Brain eQTL Almanac (Braineac) databases and Blood expression quantitative trait loci (eQTL) browser. The significant association was only found between genotype at rs1143633 and IL1B expression level of putamen and white matter in Braineac database, which was more prominent with homozygous (GG) carriers.
Conclusions
Our study firstly reported the association of IL1B polymorphism with the risk of ET in Chinese population. However, the association might only suggest a marker of IL1B SNP associated with ET instead of the casual variant. Further studies are needed to confirm our finding.
Similar content being viewed by others
Background
ET is a clinical disorder of unknown cause characterized by tremor (postural and/or kinetic) predominantly in both upper limbs and other body parts involved occasionally [1].It is not uncommon to notice an overlap of ET with RLS (characterized by dysaesthesias usually in the calves, associated with an irresistible urge to move these limbs) in some families. Clinical surveys have shown that more than half of the ET or RLS patients report at least one first degree relative was affected with either or both of these diseases [2, 3]. Dopamine transporter (DAT) binding in the striatum was shown to be decreased in some RLS patients [4] or ET patients [5], suggesting a possible dopamine dysregulation in both diseases. Genetic factors play an important role in the pathogenesis of both ET and RLS [6,7,8,9,10,11].Genome wide association study (GWAS) showed that several single nucleotide polymorphisms (SNPs) were associated with the risk of RLS, including MEIS1, BTBD9, PTPRD, MAP2K5, TOX3 and Intergenic region of 2p14 [12,13,14,15,16]. Recently, we tested those SNPs in Chinese ET and RLS patients and interestingly found a haplotype of MAP2K5/SKOR1 was both associated with ET and RLS, suggesting a possible genetic link between RLS and ET [17, 18].
Several studies have investigated other candidate genes for RLS, including HMOX1, HMOX2, VDR, IL17A, IL1B, NOS1, ADH1B and GABRR3. Garcia-Martin et al. found HMOX1 rs2071746 was associated with the risk of RLS in the Spanish population [19]. The fact that HMOX2 rs4786504C allele contributes to high-altitude adaption in Tibetans by leading to a more efficient breakdown of heme also makes us wonder if HMOX2 could be a risk factor for Chinese RLS [20]. In addition, VDR rs731236 SNP has been found associated with RLS [21]. Previous association of IL1B rs1143643, rs1143634 or rs1143633 and IL17A rs8193036 has been reported in RLS patients with HIV infection [22]. Jimenez-Jimenez et al. analyzed the possible relationship of two common SNPs (rs6413413 and rs1229984) in ADH1B with the risk for RLS or Parkinson’s disease and found that rs1229984 SNP was associated with the risk for not only RLS [23], but also with the risk for Parkinson’s disease in women [24]. Furthermore, two SNPs (rs7977109 and rs693534) in NOS1 have been found associated with German RLS, and only rs7977109 remained significant after correction of multiple testing, while both SNPs were not related to RLS in Spanish population [25, 26]. A recent report described an association between GABRR3 and the risk for RLS [27]. Among these possible candidate genes for RLS, HMOX1 rs2071746 and HMOX2 rs1051308 polymorphisms have been found associated with ET patients in a Spanish population [28], while lack of association of GABRR3 [29] or ADH2 [30] with ET was described in previous studies.
Therefore, in this study, we selected 12 SNPs (HMOX1 rs2071746, HMOX2 rs4786504, HMOX2 rs1051308, VDR rs731236, IL17A rs8193036, IL1B rs1143643, IL1B rs1143634, IL1B rs1143633, NOS1 rs693534, NOS1 rs7977109, ADH1B rs6413413 and ADH1B rs1229984) within seven suspected RLS risk genetic loci (HMOX1, HMOX2, VDR, IL17A, IL1B, NOS1 and ADH1B), which had not been tested in Chinese ET patients yet, to further investigate the relationship of these genetic risk factors with ET in Chinese population.
Methods
Study population
Two movement disorder specialists independently examined the suspected ET cases by using the standardized tremor examination described by Louis et al. [31, 32] and a full neurological examination was also performed to exclude Parkinsonism and other movement disorders. When there was disagreement between the two movement disorder specialists, a senior movement disorder specialist would evaluate the individuals and make the final diagnosis. The clinical diagnosis of ET was in accordance with the Consensus Statement of the Movement Disorders Society in 1998 [33]. Participants with parkinsonism, drug induced tremors, cerebellar tremor, dystonia and tremors with hyperthyroidism were all excluded from the study. As it is not uncommon for co-morbidity of RLS with ET, an RLS specialist interviewed all those ET patients, and diagnosis of RLS was made based on revised International RLS Study Group diagnostic criteria [34]. Control subjects were recruited through community population and evaluated by RLS and movement disorder specialists for exclusion of ET and RLS. All participants signed consent forms and this study was approved by the ethic committee of Ruijin Hospital affiliated to Shanghai Jiao Tong University School of Medicine.
DNA preparations and genotyping
About 3 mL blood samples were collected from ET patients and controls. DNA was extracted using phenol-chloroform-isopropyl alcohol method [35]. PCR and extension primers were designed using Primer5 software. SeqMan software was used to identify rs4786504, rs6413413 and rs1229984. Detail of primers and reaction condition of those 3 SNPs were described (See, Additional file 1: Tables S1-S3). Genotyping of other 9 variants was performed using Multiplex SNaPshot. The PCR products were purified by phosphorylase (FastAP) and exonuclease I (EXO I) and extended with ABI SNaPshot Multiplex kit. The elongated product was purified by phosphorylase (FastAP) and loaded on ABI3730xl. SNP typing was analyzed using GeneMapper 4.0 (Applied Biosystems). Detail of primers and reaction condition of those 9 SNPs were also described (See Additional file 1: Tables S4S-6).
Statistical analysis
Statistical analysis was performed using SAS (version 9.4 TS1M2; SAS Institute Inc., Cary, NC) software package. Student t test was used to compare the differences of age between ET sufferers and non-ET/RLS controls. Chi-Square test was used to compare the differences in the gender proportions and test the Hardy-Weinberg equilibrium (HWE) in the total cohort. Alleles of all SNPs distribution between ET patients and controls were also tested by Chi-square test. Risk analysis was performed by logistic regression and odds ratio (OR) was calculated with 95% confidence intervals (CI) for each SNP according to dominant or recessive models, after adjusting for age and sex. Dominant and recessive models were performed based on the assumption of an autosomal-dominant or autosomal-recessive mode of inheritance with incomplete penetrance. Multiplicative terms between age/sex and each SNP were also included in the logistic regression model. Haploview software was used to test linkage disequilibrium (LD) of SNPs in the same chromosome (Additional file 2). Haplotype analysis was also tested by haploview. R (version 3.3.1) was used to correct p values by Bonferroni method. Genetic power was calculated by Power and Sample Size Calculations (version 3.1.2). Expression quantitative loci were examined in three different databases: GTEx [36], Blood eQTL browser [37] and Braineac databases [38].
Results
Totally, 225 ET patients and 229 normal controls were enrolled in this study. Demographic feature of our samples showed that there are no significant differences of age and gender between ET patients and controls. One hundred of total recruited ET (44.44%) had a positive family history for ET. 25 ET patients (11.11%) were also diagnosed with RLS (Additional file 3).
In the first place, we excluded those 25 ET patients also diagnosed with RLS in order to exclude the effect of RLS comorbidity on the association study of SNPs with ET. No significant difference of age and gender was found between 200 ET patients without RLS and 229 controls (Table 1). Among 12 SNPs, all were in HWE except for two SNPs, rs4786504 of HMOX2 and rs6413413 of ADH1B (all were rs6413413AA genotype). Call rates of those 10 SNPs were all > 95%. After adjusting for age and gender, two SNPs of IL1B were shown associated with the risk of ET in our cohort (rs1143643: OR = 1.86, p = 0.017, recessive model; rs1143633: OR = 2.57, p = 0.003, recessive model). However, only rs1143633 of IL1B in a recessive model remained statistical significant after correcting p value by Bonferroni method (pcorrected = 0.024) (Table 2). The effect of age and sex, and their interaction with the studied SNPs in the logistic regression analysis were shown in Additional file 4. We only found significant interactions of sex with NOS1 rs693534 and ADH1B rs1229984 in the dominant model and interaction between sex and HMOX2 rs1051308 in the recessive model. In order to further exclude gender confounders, we performed a subgroup analysis and found that the association between rs1143633 of IL1B and ET remained significant (Additional file 5). No significant difference of age between ET patients and controls were found among female subgroup and male subgroup (Additional file 6).
Furthermore, we determined whether the statistical significance of IL1B still remained when ET patients with concomitant RLS were included. Among 12 SNPs, only rs1143633 of IL1B in a recessive model remained statistical significant after Bonferroni correction (OR = 2.63, p = 0.002), which was consistent with our former result (Additional file 7). The distribution of rs1143633 genotypes and allelic variants in ET patients with or without concomitant RLS and controls are shown in Table 3. The haplotype analysis showed that no haplotype block in IL1B and NOS1 was associated with ET patients (Additional file 8). The effect of age and sex, and their interaction with each SNP in the logistic regression model were similar to when ET patients with concomitant RLS were excluded (Additional file 9). In the further subgroup analysis (female/male cohort), there was no significant difference of age between ET patients and controls among female cohort and male cohort (Additional file 6). The association between rs1143633 of IL1B and ET was only shown significant in female cohort (p = 0.016) instead of male cohort (p = 0.052), after adjustment for age (Additional file 5).
Last, we performed a query in GTEx, Braineac and Blood eQTL browser to determine any phenotypic interest of rs1143633. Brain eQTL data were collected through GTEx and Braineac databases. No significant association between genotypes at rs1143633 and brain IL1B expression level was found in GTEx, while in Braineac database the G allele increased the expression in putamen (p = 8.70 × 10− 3, with probe set 2,571,522) and white matter (p = 8.50 × 10− 4, with probe set 2,571,524), and a more prominent increase were seen in homozygous carriers (Additional file 10). In blood, we did not observe a cis-eQTr trans-eQTL effect for this SNP on IL1B.
Discussion
This is the first study demonstrating a possible association of IL1B rs1143633 with ET in Chinese population, among 12 SNPs harbored in seven possible RLS genes loci, including HMOX1, HMOX2, VDR, IL17A, IL1B, NOS1 and ADH1B. In further subgroup analysis to exclude gender confounders, the significance still remained in both male and female cohort when only ET patients without RLS were included.
IL1B gene encodes for the proinflammatory cytokine interleukin-1β (IL-1β), which belongs to member of IL-1 family and is produced by several cell types including blood monocytes, tissue macrophages and cells of the central nervous system [39]. It has been found mediating chronic inflammation in neurodegenerative conditions, such as Parkinson’s disease [40, 41], Amyotrophic lateral sclerosis [42], Alzheimer’s disease [43, 44] and neurodegenerative process in inflammatory disease such as multiple sclerosis [45].
The possible pathophysiology of IL-1β in ET remains largely unclear. Two main pathogenetic mechanisms of ET have been proposed [46, 47]. The traditional olivary model attributes ET to a pathological pacemaker in the inferior olivary nucleus that drives tremor. Another model is cerebellar degenerative model which suggested that the degeneration of Purkinje neurons and connected neuronal populations played important role in the pathogenesis of ET. Based on cerebellar degenerative model, IL-1β might be involved in ET through an immune-based neurodegenerative pathogenesis. In line with this hypothesis, IL-1β can also cause an imbalance between the GABAergic and glutamatergic synaptic transmission at Purkinje cell synapses in experimental autoimmune encephalomyelitis (EAE) cerebellum, which are early events triggering secondary excitotoxicity and inflammatory neurodegeneration in EAE disease [48, 49]. However, we should be very cautious with this hypothesis as the association of IL-1β with ET in our study might only suggest a marker of IL1B SNP, and did not imply any inflammation pathogenesis in ET. Besides, we only found a significant association of genotype at rs1143633 with IL1B expression level in putamen and white matter, not cerebellum.
No significant differences were found either in the frequencies of genotypes or in the frequencies of the allelic variants of other SNPs harbored in possible RLS genes loci, including HMOX1, HMOX2, VDR, IL17A, IL1B, NOS1 and ADH1B. Our study did not replicate the previously reported association ofHMOX1 rs2071746 and HMOX2 rs1051308 polymorphisms with ET, which could be due to different ethnic populations or small sample of our study.
Some limitations of our study should be noticed. Firstly, the number of ET patients and controls recruited in our study is relatively small. Therefore, the power of our study is low (Table 2 and Additional file 7), which could lead to inflation of the effect size and potentially mimic the positive signal. Similar studies with big sample size or GWAS are warranted in the future to confirm our findings in Asians or other populations. Secondly, there could be population sub-stratification that was not corrected for due to using self-reported ethnicity instead of genetic ancestry. Furthermore, 9 variants in our study were genotyped by using Multiplex SNaPshot, which was less reliable compared to Sanger sequencing. Lastly, some ET might have a subclinical dopaminergic deficiency which could introduce an enrollment bias of ET in our cohorts. Although the diagnosis of ET is performed by two experienced movement disorder specialists, it would be better to perform a DAT-SCAN to exclude this possibility. Unfortunately, due to financial concerns, we did not perform DAT-SCAN in our ET patients.
Conclusion
We found a significant association of IL1B rs1143633 polymorphism in the recessive model with the risk for ET in Chinese population. However, the results should be taken with caution because segregation analysis of familial ET often suggests an autosomal dominant inheritance and there was no significant difference in the frequencies of allelic variants in our study. In addition, after a query in GTEx, Braineac and Blood eQTL browser, significant association between genotype at rs1143633 and IL1B expression level was only found in Braineac database in putamen and white matter, which was inconsistent with the main hypothesis of ET as a disorder with cerebellum involvement. This significant SNP was a marker and more likely not the casual variant, but in linkage disequilibrium with the casual variant. Furthermore, it is still too early to draw the conclusion that ET has relation with RLS from genetic point of view, since association of IL1B has only been reported in RLS patients with HIV infection. More studies are certainly needed in the future to replicate our finding and investigate the true causal variant.
Abbreviations
- Braineac:
-
Brain eQTL Almanac
- CI:
-
Confidence intervals
- DAT:
-
Dopamine transporter
- EAE:
-
Experimental autoimmune encephalomyelitis
- eQTL:
-
Expression quantitative trait loci
- ET:
-
Essential tremor
- GTEx:
-
Genotype-tissue Expression
- GWAS:
-
Genome wide association study
- HWE:
-
Hardy-Weinberg equilibrium
- IL-1β:
-
interleukin-1β
- LD:
-
Linkage disequilibrium
- OR:
-
Odds ratio
- PCR:
-
Polymerase chain reaction
- RLS:
-
Restless legs syndrome
- SNP:
-
Single nucleotide polymorphism
References
Jankovic J. Essential tremor: a heterogenous disorder. Mov Disord. 2002;17(4):638–44. https://doi.org/10.1002/mds.10221.
Deng H, Le W, Jankovic J. Genetics of essential tremor. Brain. 2007;130(Pt 6):1456–64. https://doi.org/10.1093/brain/awm018.
Winkelmann J, Polo O, Provini F, Nevsimalova S, Kemlink D, Sonka K, et al. Genetics of restless legs syndrome (RLS): state-of-the-art and future directions. Mov Disord. 2007;22(Suppl 18):S449–58. https://doi.org/10.1002/mds.21587.
Earley CJ, Kuwabara H, Wong DF, Gamaldo C, Salas R, Brasic J, et al. The dopamine transporter is decreased in the striatum of subjects with restless legs syndrome. Sleep. 2011;34(3):341–7.
Isaias IU, Canesi M, Benti R, Gerundini P, Cilia R, Pezzoli G, et al. Striatal dopamine transporter abnormalities in patients with essential tremor. Nucl Med Commun. 2008;29(4):349–53. https://doi.org/10.1097/MNM.0b013e3282f4d307.
Gulcher JR, Jonsson P, Kong A, Kristjansson K, Frigge ML, Karason A, et al. Mapping of a familial essential tremor gene, FET1, to chromosome 3q13. Nat Genet. 1997;17(1):84–7. https://doi.org/10.1038/ng0997-84.
Higgins JJ, Pho LT. Nee LE. A gene (ETM) for essential tremor maps to chromosome 2p22-p25. Mov Disord. 1997;12(6):859–64. https://doi.org/10.1002/mds.870120605.
Shatunov A, Sambuughin N, Jankovic J, Elble R, Lee HS, Singleton AB, et al. Genomewide scans in north American families reveal genetic linkage of essential tremor to a region on chromosome 6p23. Brain. 2006;129(Pt 9):2318–31. https://doi.org/10.1093/brain/awl120.
Stefansson H, Steinberg S, Petursson H, Gustafsson O, Gudjonsdottir IH, Jonsdottir GA, et al. Variant in the sequence of the LINGO1 gene confers risk of essential tremor. Nat Genet. 2009;41(3):277–9. https://doi.org/10.1038/ng.299.
Jimenez-Jimenez FJ, Alonso-Navarro H, Garcia-Martin E, Agundez JAG. Genetics of restless legs syndrome: an update. Sleep Med Rev. 2018;39:108–21. https://doi.org/10.1016/j.smrv.2017.08.002.
Muller SH, Girard SL, Hopfner F, Merner ND, Bourassa CV, Lorenz D, et al. Genome-wide association study in essential tremor identifies three new loci. Brain. 2016;139(Pt 12):3163–9. https://doi.org/10.1093/brain/aww242.
Winkelmann J, Czamara D, Schormair B, Knauf F, Schulte EC, Trenkwalder C, et al. Genome-wide association study identifies novel restless legs syndrome susceptibility loci on 2p14 and 16q12.1. PLoS Genet. 2011;7(7):e1002171. https://doi.org/10.1371/journal.pgen.1002171.
Winkelmann J, Schormair B, Lichtner P, Ripke S, Xiong L, Jalilzadeh S, et al. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat Genet. 2007;39(8):1000–6. https://doi.org/10.1038/ng2099.
Stefansson H, Rye DB, Hicks A, Petursson H, Ingason A, Thorgeirsson TE, et al. A genetic risk factor for periodic limb movements in sleep. N Engl J Med. 2007;357(7):639–47. https://doi.org/10.1056/NEJMoa072743.
Schormair B, Kemlink D, Roeske D, Eckstein G, Xiong L, Lichtner P, et al. PTPRD (protein tyrosine phosphatase receptor type delta) is associated with restless legs syndrome. Nat Genet. 2008;40(8):946–8. https://doi.org/10.1038/ng.190.
Fuh JL, Chung MY, Yao SC, Chen PK, Liao YC, Hsu CL, et al. Susceptible genes of restless legs syndrome in migraine. Cephalalgia. 2016;36(11):1028–37. https://doi.org/10.1177/0333102415620907.
Chen J, Huang P, He Y, Shen J, Du J, Cui S, et al. A haplotype of MAP2K5/SKOR1 was associated with essential tremor in Chinese population. Parkinsonism Relat Disord. 2018;53:118–9. https://doi.org/10.1016/j.parkreldis.2018.05.017.
Li G, Tang H, Wang C, Qi X, Chen J, Chen S, et al. Association of BTBD9 and MAP2K5/SKOR1 with restless legs syndrome in Chinese population. Sleep. 2017;40:4. https://doi.org/10.1093/sleep/zsx028.
Garcia-Martin E, Jimenez-Jimenez FJ, Alonso-Navarro H, Martinez C, Zurdo M, Turpin-Fenoll L, et al. Heme Oxygenase-1 and 2 common genetic variants and risk for restless legs syndrome. Medicine. 2015;94(34):e1448. https://doi.org/10.1097/md.0000000000001448.
Yang D, Peng Y, Ouzhuluobu, Bianbazhuoma, Cui C, Bianba, et al. HMOX2 functions as a modifier gene for high-altitude adaptation in Tibetans. Hum Mutat. 2016;37(2):216–23. https://doi.org/10.1002/humu.22935.
Jimenez-Jimenez FJ, Garcia-Martin E, Alonso-Navarro H, Martinez C, Zurdo M, Turpin-Fenoll L, et al. Association between vitamin D receptor rs731236 (Taq1) polymorphism and risk for restless legs syndrome in the Spanish Caucasian population. Medicine. 2015;94(47):e2125. https://doi.org/10.1097/md.0000000000002125.
Hennessy MD, Zak RS, Gay CL, Pullinger CR, Lee KA, Aouizerat BE. Polymorphisms of interleukin-1 Beta and interleukin-17Alpha genes are associated with restless legs syndrome. Biol Res Nurs. 2014;16(2):143–51. https://doi.org/10.1177/1099800413478827.
Jimenez-Jimenez FJ, Gomez-Tabales J, Alonso-Navarro H, Zurdo M, Turpin-Fenoll L, Millan-Pascual J, et al. Association between the rs1229984 polymorphism in the alcohol dehydrogenase 1B gene and Risk for restless legs syndrome. Sleep. 2017;40(12). https://doi.org/10.1093/sleep/zsx174.
Garcia-Martin E, Diez-Fairen M, Pastor P, Gomez-Tabales J, Alonso-Navarro H, Alvarez I, et al. Association between the missense alcohol dehydrogenase rs1229984T variant with the risk for Parkinson's disease in women; 2018. https://doi.org/10.1007/s00415-018-9136-9.
Winkelmann J, Lichtner P, Schormair B, Uhr M, Hauk S, Stiasny-Kolster K, et al. Variants in the neuronal nitric oxide synthase (nNOS, NOS1) gene are associated with restless legs syndrome. Mov Disord. 2008;23(3):350–8. https://doi.org/10.1002/mds.21647.
Jimenez-Jimenez FJ, Alonso-Navarro H, Martinez C, Zurdo M, Turpin-Fenoll L, Millan-Pascual J, et al. Neuronal nitric oxide synthase (nNOS, NOS1) rs693534 and rs7977109 variants and risk for restless legs syndrome. J Neural Transmi (Vienna, Austria : 1996). 2015;122(6):819–23. https://doi.org/10.1007/s00702-014-1322-z.
Jimenez-Jimenez FJ, Esguevillas G, Alonso-Navarro H, Zurdo M, Turpin-Fenoll L, Millan-Pascual J, et al. Gamma-aminobutyric acid (GABA) receptors genes polymorphisms and risk for restless legs syndrome. Pharmacogenomics J. 2018;18(4):565–77. https://doi.org/10.1038/s41397-018-0023-7.
Ayuso P, Agundez JA, Alonso-Navarro H, Martinez C, Benito-Leon J, Ortega-Cubero S, et al. Heme oxygenase 1 and 2 common genetic variants and risk for essential tremor. Medicine. 2015;94(24):e968. https://doi.org/10.1097/md.0000000000000968.
Garcia-Martin E, Martinez C, Alonso-Navarro H, Benito-Leon J, Lorenzo-Betancor O, Pastor P, et al. Gamma-aminobutyric acid (GABA) receptor rho (GABRR) polymorphisms and risk for essential tremor. J Neurol. 2011;258(2):203–11. https://doi.org/10.1007/s00415-010-5708-z.
Martinez C, Garcia-Martin E, Alonso-Navarro H, Benito-Leon J, Puertas I, Rubio L, et al. Alcohol dehydrogenase 2 genotype and allelic variants are not associated with the risk for essential tremor. Clin Neuropharmacol. 2007;30(4):196–200. https://doi.org/10.1097/wnf.0b013e3180413d94.
Louis ED, Ferreira JJ. How common is the most common adult movement disorder? Update on the worldwide prevalence of essential tremor. Mov Disord. 2010;25(5):534–41. https://doi.org/10.1002/mds.22838.
Louis ED, Ford B, Frucht S, Barnes LF, M XT, Ottman R. Risk of tremor and impairment from tremor in relatives of patients with essential tremor: a community-based family study. Ann Neurol. 2001;49(6):761–9.
Deuschl G, Bain P, Brin M. Consensus statement of the Movement Disorder Society on Tremor. Ad Hoc Sci Comm Mov Disord. 1998;13(Suppl 3):2–23.
Allen RP, Picchietti DL, Garcia-Borreguero D, Ondo WG, Walters AS, Winkelman JW, et al. Restless legs syndrome/Willis-Ekbom disease diagnostic criteria: updated international restless legs syndrome study group (IRLSSG) consensus criteria--history, rationale, description, and significance. Sleep Med. 2014;15(8):860–73. https://doi.org/10.1016/j.sleep.2014.03.025.
Ahmad NN, Cu-Unjieng AB, Donoso LA. Modification of standard proteinase K/phenol method for DNA isolation to improve yield and purity from frozen blood. J Med Genet. 1995;32(2):129–30.
The Genotype-Tissue Expression (GTEx) project. Nature genetics. 2013;45 6:580–5; doi: https://doi.org/10.1038/ng.2653.
Westra H-J, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238. https://doi.org/10.1038/ng.2756.
Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17(10):1418–28. https://doi.org/10.1038/nn.3801.
Dinarello CA. Biology of interleukin 1. FASEB J. 1988;2(2):108–15.
Calabrese V, Santoro A, Monti D, Crupi R, Di Paola R, Latteri S, et al. Aging and Parkinson's disease: Inflammaging, neuroinflammation and biological remodeling as key factors in pathogenesis. Free Radic Biol Med. 2018;115:80–91. https://doi.org/10.1016/j.freeradbiomed.2017.10.379.
Sampaio TB, Marcondes Sari MH, Pesarico AP, Mantovani AC, Zeni G, Nogueira CW. 7-Fluoro-1,3-diphenylisoquinoline reverses motor and non-motor symptoms induced by MPTP in mice: role of striatal neuroinflammation. Eur J Pharmacol. 2018;819:129–35. https://doi.org/10.1016/j.ejphar.2017.12.001.
Saberi D, Ott B, Dahlke C, Matschke V, Schmitt-John T, Theiss C. The spatiotemporal pattern of degeneration in the cerebellum of the wobbler mouse. J Neuropathol Exp Neurol. 2016;75(4):347–57. https://doi.org/10.1093/jnen/nlw005.
Sawikr Y, Yarla NS, Peluso I, Kamal MA, Aliev G, Bishayee A. Neuroinflammation in Alzheimer's disease: the preventive and therapeutic potential of polyphenolic nutraceuticals. Adv Protein Chem Struct Biol. 2017;108:33–57. https://doi.org/10.1016/bs.apcsb.2017.02.001.
Frost GR, Li YM. The role of astrocytes in amyloid production and Alzheimer's disease. 2017;7 12; doi: https://doi.org/10.1098/rsob.170228.
Koudriavtseva T, Mainero C. Neuroinflammation, neurodegeneration and regeneration in multiple sclerosis: intercorrelated manifestations of the immune response. Neural Regen Res. 2016;11(11):1727–30. https://doi.org/10.4103/1673-5374.194804.
Louis ED. Re-thinking the biology of essential tremor: from models to morphology. Parkinsonism Relat Disord. 2014;20(Suppl 1):S88–93. https://doi.org/10.1016/s1353-8020(13)70023-3.
Louis ED. Understanding essential tremor: progress on the biological front. Curr Neurol Neurosci Rep. 2014;14(6):450. https://doi.org/10.1007/s11910-014-0450-z.
Mandolesi G, Gentile A, Musella A, Centonze D. IL-1beta dependent cerebellar synaptopathy in a mouse mode of multiple sclerosis. Cerebellum (London, England). 2015;14(1):19–22. https://doi.org/10.1007/s12311-014-0613-0.
Mandolesi G, Musella A, Gentile A, Grasselli G, Haji N, Sepman H, et al. Interleukin-1beta alters glutamate transmission at purkinje cell synapses in a mouse model of multiple sclerosis. J Neurosci. 2013;33(29):12105–21. https://doi.org/10.1523/jneurosci.5369-12.2013.
Acknowledgements
We would like to thank all subjects participating in this study.
Funding
This study was supported by grants from National Natural Science Fund (81571103, 81771374), the National Key R&D Program of China (2016YFC1306000).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
JC and PH collected ET and control data, performed the statistical analysis and drafted the manuscript. YCH, JYS, JJD and SSC helped to collect ET data. SDC supervised the study and revised the manuscript. JFM designed the study and revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Institutional Ethics Committee of Shanghai Jiao Tong University School of Medicine. Written consent form was obtained from each participant.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Detail of primers and reaction condition of 12 selected SNPs. (DOCX 15 kb)
Additional file 2:
linkage disequilibrium (LD) of SNPs in the same chromosome. (DOCX 13 kb)
Additional file 3:
Demographic Information of Cases (all ET patients) and Controls (DOCX 12 kb)
Additional file 4:
The effect of age and sex, and their interaction with each SNP (ET without RLS) (DOCX 19 kb)
Additional file 5:
Subgroup analysis of association between rs1143633 and ET risk in recessive model (DOCX 15 kb)
Additional file 6:
Comparison of age between ET patients and controls in subgroup analysis (DOCX 13 kb)
Additional file 7:
Association of SNPs of candidate genes and odds ratio to ET risk (all ET patients) (DOCX 17 kb)
Additional file 8:
Haplotype association study of IL1B and NOS1 (DOCX 13 kb)
Additional file 9:
The effect of age and sex, and their interaction with each SNP (all ET patients) (DOCX 19 kb)
Additional file 10:
Effects of rs1143633 genotype on brain IL1B expression level. (DOCX 28 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Chen, J., Huang, P., He, Y. et al. IL1B polymorphism is associated with essential tremor in Chinese population. BMC Neurol 19, 99 (2019). https://doi.org/10.1186/s12883-019-1331-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12883-019-1331-5