Abstract
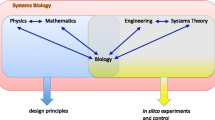
The progress of the molecular biosciences has been so enormous that a discipline studying how cellular functioning emerges out of the behaviors of their molecular constituents has become reality. Systems biology studies cells as spatiotemporal networks of interacting molecules using an integrative approach of theory (mathematics, physics, engineering), experimental biology (genetics, molecular biology, physiology), and quantitative network-wide analytical measurement (analytical biochemistry, imaging). Its aim is to understand how molecules jointly bring about life. Systems biology is rapidly discovering principles governing the functioning of molecular networks and methods to model and measure them. The application of the conceptual strength of systems sciences such as physics and engineering are opening up new ways of experimentally studying the design and functioning of molecular networks. Typically, in such studies models are used to highlight principles, design experiments, or to act in a predictive mode to specifically modify cellular behavior. In this trend article, theoretical approaches being applied in systems biology will be discussed. A number of books have recently appeared that deal with theoretical aspects of systems biology. In this article they are highlighted to facilitate the communication of systems biology to a broader audience.
Similar content being viewed by others
References
Acar M, Becskei A, van Oudenaarden A (2005) Enhancement of cellular memory by reducing stochastic transitions. Nature 435: 228–232.
Albert R, Barabasi AL (2002) Statistical mechanics of complex networks. Reviews of Modern Physics 74: 47–97.
Almaas E, Kovács B, Vicsek T, Oltvai ZN, Barabasi A-L (2004) Global organization of metabolic fluxes in the bacterium Escherichia coli. Nature 427: 839–843.
Alon U (2006) Introduction to Systems Biology. London: Chapman and Hall.
Bakker BM, Michels PA, Opperdoes FR, Westerhoff WV (1997) Glycolysis in bloodstream form Trypanosoma brucei can be understood in terms of the kinetics of the glycolytic enzymes. Journal of Biological Chemistry 272: 3207–3215.
Barabasi AL, Oltvai ZN (2004) Network biology: Understanding the cell’s functional organization. Nature Reviews Genetics 5: 101–113.
Benner SA, Sismour AM (2005) Synthetic biology. Nature Reviews Genetics 6: 533–543.
Boogerd FC, Bruggeman FJ, Hofmeyr J-H, Westerhoff HV, eds. (2007) Systems Biology: Philosophical Foundations. Amsterdam: Elsevier Science.
Bruggeman FJ, Boogerd FC, Westerhoff HV (2005) The multifarious short-term regulation of ammonium assimilation of Escherichia coli: Dissection using an in silico replica. FEBS Journal 272: 1965–1985.
Bruggeman FJ, de Haan J, Hardin H, Bouwman J, Rossell S, van Eunen K, Bakker BM, Westerhoff HV (2006) Time-dependent hierarchical regulation analysis: Deciphering cellular adaptation. Systems Biology (Stevenage) 153: 318–322.
Bruggeman FJ, Westerhoff HV (2007) The nature of systems biology. Trends in Microbiology 15: 45–50.
Bruggeman FJ, Westerhoff HV, Boogerd FC (2002) BioComplexity: A pluralist research strategy is necessary for a mechanistic explanation of the “live” state. Philosophical Psychology 15: 411–140.
Csete M, Doyle J (2004) Bow ties, metabolism and disease. Trends in Biotechnology 22: 446–50.
Danø S, Sørensen PG, Hynne F (1999) Sustained oscillations in living cells. Nature 402: 320–322.
El-Samad H, Kurata H, Doyle JC, Gross CA, Khammash M (2005) Surviving heat shock: Control strategies for robustness and performance. Proceedings of the National Academy of Sciences USA 102: 2736–2741.
Fell DA (1997) Understanding the Control of Metabolism. Miami: Portland Press.
Galperin MY (2005) A census of membrane-bound and intracellular signal transduction proteins in bacteria: Bacterial IQ, extroverts and introverts. BMC Microbiology 5: 35.
Goodacre R, Vaidyanathan S, Dunn WB, Harrigan GG, Kell DB (2004) Metabolomics by numbers: Acquiring and understanding global metabolite data. Trends in Biotechnology 22: 245–252.
Heinrich R, Rapoport TA (1974) A linear steady-state treatment of enzymatic chains: General properties, control and effector strength. European Journal of Biochemistry 42: 89–95.
Hoefnagel MH, Starrenburg MJ, Martens DE, Hugenholtz J, Kleerebezem M, Van Swam II, Bongers R, Westerhoff HV, Snoep JL (2002) Metabolic engineering of lactic acid bacteria, the combined approach: Kinetic modelling, metabolic control and experimental analysis. Microbiology 148(Pt 4): 1003–1013.
Hoops S, Sahle S, Lee C, Pahle J, Simus N, Singhal M, Xu L, Mendes P, Kummer U (2006) COPASI—a COmplex PAthway SImulator. Bioinformatics 22: 3067–3074.
Hornberg JJ, Bruggeman FJ, Binder B, Geest CR, Bij de Vaatel AJM, Lankelma J, Heinrich JR, Westerhoff HV (2005) Principles behind the multifarious control of signal transduction: ERK phosphorylation and kinase/phosphatase control. FEBS Journal 272: 244–258.
Hucka M, Finney A, Sauro HM, Bolouri H, Doyle J, Kitano H (2002) The ERATO Systems Biology Workbench: Enabling interaction and exchange between software tools for computational biology. Pacific Symposium on Biocomputation: 450–461.
Hynne R, Danø S, Stensen PG (2001) Full-scale model of glycolysis in Saccharomyces cerevisiae. Biophysical Chemistry 94: 121–163.
Kacser H, Burns JA (1973) The control of flux. Symposia of the Society for Experimental Biology 27: 65–104.
Kalir S, Mangan S, Alon U (2005) A coherent feed-forward loop with a SUM input function prolongs flagella expression in Escherichia coli. Molecular Systems Biology 1: 2005: 0006.
Kaneko K (2006) Life: An Introduction to Complex Systems Biology. Berlin: Springer.
Kaufmann BB, van Oudenaarden A (2007) Stochastic gene expression: From single molecules to the proteome. Current Opinion in Genetics and Development 17: 107–112.
Kholodenko BN, Demin OV, Moehren G, Hoek JB (1999) Quantification of short term signaling by the epidermal growth factor receptor. Journal of Biological Chemistry 274: 30169–30181.
Klipp E, Herwig R, Kowald A, Wierling C, Lehrach H (2005) Systems Biology in Practice: Concepts, Implementation, and Application. Weinheim: Wiley-VCH.
Klipp E, Nordlander B, Kruger R, Gennemark P, Hohmann S (2005) Integrative model of the response of yeast to osmotic shock. Nature Biotechnology 23: 975–982.
Mahadevan R, Schilling CH (2003) The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metabolic Engineering 5: 264–276.
Mangan S, Alon U (2003) Structure and function of the feed-forward loop network motif. Proceedings of the National Academy of Sciences USA 100: 11980–11985.
Mangan S, Itzkovitz S, Zaslaver A, Alon U (2006) The incoherent feedforward loop accelerates the response-time of the gal system of Escherichia coli. Journal of Molecular Biology 356: 1073–1081.
Mangan S, Zaslaver A, Alon U (2003) The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. Journal of Molecular Biology 334: 197–204.
Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U (2002) Network motifs: Simple building blocks of complex networks. Science 298: 824–827.
Moles CG, Mendes P, Banga JR (2003) Parameter estimation in biochemical pathways: A comparison of global optimization methods. Genome Research 13: 2467–2474.
Mushegian AR, Koonin EV (1996) A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proceedings of the National Academy of Sciences USA 93: 10268–10273.
Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG, Edwards SW, McDowell HP, Unitt JF, Sullivan E, Grimley R, Benson N, Broomhead D, Kell DB, White MRH (2004) Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science 306: 704–708.
Newman MEJ (2003) The structure and function of complex networks. SIAM Review 45: 167–256.
Olivier BG, Snoep JL (2004) Web-based kinetic modelling using JWS Online. Bioinformatics 20: 2143–2144.
Ozbudak EM, Thattai M, Lim HN, Shraiman BI, van Oudenaarden A (2004) Multistability in the lactose utilization network of Escherichia coli. Nature 427: 737–740.
Palsson BO (2006) Systems Biology: Properties of Reconstructed Networks. Cambridge: Cambridge University Press.
Papin JA, Stelling J, Price ND, Klamt S, Schuster S, Palsson BO (2004) Comparison of network-based pathway analysis methods. Trends in Biotechnology 22: 400–405.
Paulsson J (2004) Summing up the noise in gene networks. Nature 427: 415–418.
Price ND, Reed JL, Palsson BO (2004) Genome-scale models of microbial cells: Evaluating the consequences of constraints. Nature Reviews Microbiology 2: 886–897.
Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi A-L (2002) Hierarchical organization of modularity in metabolic networks. Science 297: 1551–1555.
Reed JL, Palsson BO (2004) Genome-scale in silico models of E. coli have multiple equivalent phenotypic states: Assessment of correlated reaction subsets that comprise network states. Genome Research 14: 1797–1805.
Reijenga KA, Bakker BM, van der Weijden CC, Westerhoff HV (2005) Training of yeast cell dynamics. FEBS Journal 272: 1616–1624.
Rossell S, Van der Weijden CC, Lindenbergh A, van Tuijl A, Francke C, Bakker BM, Westerhoff HV (2006) Unraveling the complexity of flux regulation: A new method demonstrated for nutrient starvation in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences USA 103: 2166–2171.
Schoeberl B, Eichler-Jonsson C, Gilles ED, Müller G (2002) Computational modeling of the dynamics of the MAP kinase cascade activated by surface and internalized EGF receptors. Nature Biotechnology 20: 370–375.
Segel IH (1993) Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-state Enzyme Systems. New York: Wiley.
Snoep JL, Bruggeman F, Olivier BG, Westerhoff HV (2006) Towards building the silicon cell: A modular approach. Biosystems 83: 207–216.
Sprinzak D, Elowitz MB (2005) Reconstruction of genetic circuits. Nature 438: 443–448.
Stelling J, Klamt S, Bettenbrock K. Schuster S, Gilles ED (2002) Metabolic network structure determines key aspects of functionality and regulation. Nature 420: 190–193.
Swameye I, Muller TG, Timmer J, Sandra O, Klingmuller U (2003) Identification of nucleocytoplasmic cycling as a remote sensor in cellular signaling by databased modeling. Proceedings of the National Academy of Sciences USA 100: 1028–1033.
Szallazi Z, Stelling J, Periwal V (2006) System Modeling in Cellular Biology: From Concepts to Nuts and Bolts. Cambridge, MA: MIT Press.
Szathmáry E (2004) From biological analysis to synthetic biology. Current Biology 14: R145–R146.
Ter Kuile BH, Westerhoff HV (2001) Transcriptomemeets metabolome: Hierarchical and metabolic regulation of the glycolytic pathway. FEBS Letters 500: 169–171.
Teusink B, Passarge J, Reijenga CA, Esgalhado E, van der Weijden CC, Schepper M, Walsh MC, Bakker BM, van Dam K, Westerhoff HV, Snoep JL (2000) Can yeast glycolysis be understood in terms of in vitro kinetics of the constituent enzymes? Testing biochemistry. European Journal of Biochemistry 267: 5313–5329.
Tyson JJ, Chen K, Novak B (2001) Network dynamics and cell physiology. Nature Reviews Molecular Cell Biology 2: 908–916.
Westerhoff HV, Chen YD (1984) How do enzyme activities control metabolite concentrations? An additional theorem in the theory of metabolic control. European Journal of Biochemistry 142: 425–430.
Westerhoff HV, Palsson BO (2004) The evolution of molecular biology into systems biology. Nature Biotechnology 22: 1249–1252.
Wiechert W (2002) An introduction to 13C metabolic flux analysis. Genetic Engineering 24: 215–238.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bruggeman, F.J. Systems Biology: At Last an Integrative Wet and Dry Biology!. Biol Theory 2, 183–188 (2007). https://doi.org/10.1162/biot.2007.2.2.183
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1162/biot.2007.2.2.183