Abstract
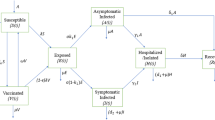
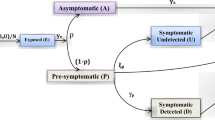
The global emergence of COVID-19 and its widespread transmission posed a formidable challenge for the global medical community. While vaccinations succeeded in mitigating the severity and fatality of the infection, a new challenge emerged: addressing transmission in the presence of comorbidities. A comprehensive mathematical model has been developed to address this issue, incorporating elements such as nonpharmaceutical interventions, vaccination strategies, comorbidity factors, limited healthcare resources, and the impact of nosocomial transmission. This updated model is formulated as a set of nonlinear partial differential equations under the category of reaction-diffusion models, aiming to provide a more accurate representation of the dynamics and interactions involved in spreading infectious diseases in a given population. The methodology employed involves a comprehensive analysis of the master model system’s qualitative characteristics, focussing on the stability of its constituent subsystems. The model’s dynamical system is subjected to numerical solutions, enabling a detailed exploration of its behaviour under various conditions. A rigorous parametric variation is carried out to understand the model’s response to different parameter values. The novelty of this research is rooted in its pioneering approach to bridging the gap between theory and real-world observations. By rigorously validating theoretical results against empirical experimental data, the research aims to provide valuable insights into the dynamics of the ongoing pandemic. The outcomes generated by the present model system are expected to offer a deeper and more comprehensive understanding of the pandemic’s behaviour and transmission patterns, playing a pivotal role in advancing the field of theoretical modelling.




































Similar content being viewed by others
Data Availability Statement
Data sharing does not apply to this article as no dataset and code were generated or analysed during the present study.
References
J.T. Wu, K. Leung, G.M. Leung, Nowcasting and forecasting the potential domestic and international spread of the 2019-NCOV outbreak originating in wuhan, china: a modelling study. Lancet. 395(10225), 689–697 (2020). https://doi.org/10.1016/S0140-6736(20)30260-9
G.Q. Sun, S.F. Wang, M.T. Li, L. Li, J. Zhang, W. Zhang, Z. Jin, G.L. Feng, Transmission dynamics of Covid-19 in Wuhan, China: effects of lockdown and medical resources. Nonlinear Dyn. 101(3), 1981–1993 (2020). https://doi.org/10.1007/s11071-020-05770-9
Q.Y. Lin, S. Zhao, D.Z. Gao, W.M. Wang, L. Yang, D.H. He, A conceptual model for the coronavirus disease 2019 (COVID-19) outbreak in Wuhan, China with individual reaction and governmental action. Int. J. Infect. Dis. 93, 211–216 (2020). https://doi.org/10.1016/j.ijid.2020.02.058
E. Shim, A. Tariq, W. Choi, Y. Lee, G. Chowell, Transmission potential and severity of COVID-19 in South Korea. Int. J. Infect. Dis. 93, 339–344 (2020). https://doi.org/10.1016/j.ijid.2020.03.031
C.N. Ngonghala, E. Iboi, S. Eikenberry, M. Scotch, C.R. MacIntyre, M.H. Bonds, A.B. Gumel, Mathematical assessment of the impact of non-pharmaceutical interventions on curtailing the 2019 novel Coronavirus. Math. Biosci. 325, 108364 (2020). https://doi.org/10.1016/j.mbs.2020.108364
D.S. Hui, E.I. Azhar, T.A. Madani, F. Ntoumi, R. Kock, O. Dar, G. Ippolito, T.D. Mchugh, Z.A. Memish, C. Drosten, The continuing 2019-NCOV epidemic threat of novel coronaviruses to global health-the latest 2019 novel coronavirus. Int. J. Infect. Dis. 91, 264–266 (2020). https://doi.org/10.1016/j.ijid.2020.01.009
R. Thompson, Pandemic potential of 2019-NCOV. Lancet Infect Dis. 20(3), 280 (2020). https://doi.org/10.1016/S1473-3099(20)30068-2
J. Yang, Y. Zheng, X. Gou, P. Ke, Z. Chen, Q. Guo, R. Ji, H. Wang, Y. Wang, Y. Zhou, Prevalence of comorbidities and its effects in patients infected with SARS-COV-2: a systematic review and meta-analysis. Int. J. Infect. Dis. 94, 91–5 (2020). https://doi.org/10.1016/j.ijid.2020.03.017
W.J. Guan, W.H. Liang, Comorbidity and its impact on 1590 patients with Covid-19 in China: a nationwide analysis. Eur. Respir. J. 55(5), 2000547 (2020). https://doi.org/10.1183/13993003.00547-2020
H. Carreira, H. Strongman, M. Peppa, H.I. McDonald, I. dos Santos-Silva, S. Stanway, L. Smeeth, K. Bhaskaran, Prevalence of Covid-19-related risk factors and risk of severe influenza outcomes in cancer survivors: a matched cohort study using linked english electronic health records data. E. Clin. Med. 29, 100656 (2020). https://doi.org/10.1016/j.eclinm.2020.100656
X. Liu, X. Zheng, B. Balachandran, Covid-19: data driven dynamics, statistical and distributed delay models, and observations. Nonlinear Dyn. 101(3), 1527–1543 (2020). https://doi.org/10.1007/s11071-020-05863-5
O. Khyar, K. Allali, Global dynamics of a multi-strain SEIR epidemic model with general incidence rates: application to Covid-19 pandemic. Nonlinear Dyn. 102(1), 489–509 (2020). https://doi.org/10.1007/s11071-020-05929-4
A. Ianni, N. icola Rossi, Describing the COVID-19 outbreak during the lockdown: fitting modified SIR models to data. Eur. Phys. J. Plus 135, 885 (2020). https://doi.org/10.1140/epjp/s13360-020-00895-7
S. Olaniyi, O.S. Obabiyi, K.O. Okosun, A.T. Oladipo, S.O. Adewale, Mathematical modelling and optimal cost-effective control of COVID-19 transmission dynamics. Eur. Phys. J. Plus. 135, 938 (2020). https://doi.org/10.1140/epjp/s13360-020-00954-z
T. Cheema, M. Raja, I. Ahmad, S. Naz, H. Ilyas, M. Shoaib, Intelligent computing with Levenberg-Marquardt artificial neural networks for nonlinear system of COVID-19 epidemic model for future generation disease control. Eur. Phys. J. Plus. 135, 932 (2020). https://doi.org/10.1140/epjp/s13360-020-00910-x
G. Rohith, K.B. Devika, Dynamics and control of Covid-19 pandemic with nonlinear incidence rates. Nonlinear Dyn. 101(3), 2013–2026 (2020). https://doi.org/10.1007/s11071-020-05774-5
P. Das, S. Das, R.K. Upadhyay, P. Das, Optimal treatment strategies for delayed cancer-immune system with multiple therapeutic approach. Chaos Solitons Fract. 136, 109806 (2020). https://doi.org/10.1016/j.chaos.2020.109806
J. Huang, G. Qi, Effects of control measures on the dynamics of Covid-19 and double-peak behavior in Spain. Nonlinear Dyn. 101(3), 1889–1899 (2020). https://doi.org/10.1007/s11071-020-05901-2
P. Das, S.S. Nadim, S. Das, P. Das, Dynamics of COVID-19 transmission with comorbidity: a data driven modelling based approach. Nonlinear Dyn. 106, 1197–1211 (2021). https://doi.org/10.1007/s11071-021-06324-3
Q. Li, B. Tang, N.L. Bragazzi, Y. Xiao, J. Wu, Modeling the impact of mass influenza vaccination and public health interventions on COVID-19 epidemics with limited detection capability. Math. Biosci. 325, 108378 (2020). https://doi.org/10.1016/j.mbs.2020.108378
B. Tang, X. Wang, Q. Li, N.L. Bragazzi, Y. Sanyi Tang, JWu. Xiao, Estimation of the transmission risk of the 2019-NCoV and its implication for public health interventions. J. Clin. Med. 9(2), 462 (2020). https://doi.org/10.3390/jcm9020462
P. Ghosh, R. Ghosh, B. Chakraborty, COVID-19 in India: statewise analysis and prediction. JMIR Public Health Surveill. 6(3), e20341 (2020). https://doi.org/10.2196/20341
Acknowledgements
In light of the improvement of the manuscript in its present revised version, the anonymous reviewers are gratefully acknowledged by the author for their invaluable comments and suggestions.
Funding
No funding received from any source to carry out this research work.
Author information
Authors and Affiliations
Contributions
As the article is written by single author, it is not applicable here.
Corresponding author
Ethics declarations
Conflict of interest
The author declares that she has no conflict of interests.
Ethical approval
The author has full consent to participate and publish.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chakravarty, K. The effect of vaccination on COVID-19 transmission dynamics with comorbidity using reaction–diffusion model. Eur. Phys. J. Plus 138, 1140 (2023). https://doi.org/10.1140/epjp/s13360-023-04766-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1140/epjp/s13360-023-04766-9