Abstract
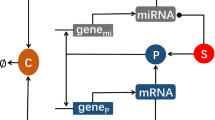
MicroRNAs are extensively known for post-transcriptional gene regulation and pattern formation in the embryonic developmental stage. We explore the origin of these spatio-temporal patterns mathematically, considering three different motifs here. For three scenarios, (1) simple microRNA-based mRNA regulation with a graded response in output, (2) microRNA-based mRNA regulation resulting in bistability in the dynamics, and (3) a coordinated response of microRNA (miRNA), simultaneously regulating the mRNAs of two different pools, detailed dynamical analysis, as well as the reaction–diffusion scenario have been considered and analyzed in the steady state and for the transient dynamics further. We have observed persistent-temporal patterns, as a result of the dynamics of the motifs, that explain spatial gradients and relevant patterns formed by related proteins in development and phenotypic heterogenetic aspects in biological systems. Competitive effects of miRNA regulation have also been found to be capable to cause spatio-temporal patterns, persistent enough to direct developmental decisions. Under coordinated regulation, miRNAs are found to generate spatio-temporal patterning even from complete homogeneity in concentration of target protein, which may have impactful insights in choice of cell fates.













Similar content being viewed by others
Data Availability
The manuscript has no associated data.
Notes
Link for the video is: https://youtu.be/C4zovgxtCpQ.
References
A. Armenta-Medina, D. Lepe-Soltero, D. Xiang, R. Datla, C. Abreu-Goodger, C.S. Gillmor, Arabidopsis thaliana miRNAs promote embryo pattern formation beginning in the zygote. Dev. Biol. 431(2), 145–151 (2017)
M. Banerjee, S. Pal, P.R. Chowdhury, Stationary and non-stationary pattern formation over fragmented habitat. Chaos Solitons Fractals 162, 112412 (2022)
I. Barbier, R. Perez-Carrasco, Y. Schaerli, Controlling spatiotemporal pattern formation in a concentration gradient with a synthetic toggle switch. Mol. Syst. Biol. 16(6), e9361 (2020)
D.P. Bartel, MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2), 281–297 (2004)
M. Bhaskaran, M. Mohan, MicroRNAs: history, biogenesis, and their evolving role in animal development and disease. Vet. Pathol. 51(4), 759–774 (2014)
M. Bianchi, A. Renzini, S. Adamo, V. Moresi, Coordinated actions of microRNAs with other epigenetic factors regulate skeletal muscle development and adaptation. Int. J. Mol. Sci. 18(4), 840 (2017)
U. Bissels, A. Bosio, W. Wagner, Micrornas are shaping the hematopoietic landscape. Haematologica 97(2), 160 (2012)
L. Borin, M. Weir, G. Schubiger, Egg ligation alter the BCD protein gradient and segmentation gene expression in embryos of drosophila. Mech. Dev. 42(1–2), 97–111 (1993)
I. Bose, S. Ghosh, Origins of binary gene expression in post-transcriptional regulation by microRNAs. Eur. Phys. J. E 35, 1–8 (2012)
G.A. Calin, C.M. Croce, Microrna signatures in human cancers. Nat. Rev. Cancer 6(11), 857–866 (2006)
P. Chakraborty, S. Ghosh, Emergent correlations in gene expression dynamics as footprints of resource competition. Eur. Phys. J. E 44, 1–12 (2021)
P. Chakraborty and S. Ghosh. Emergent regulatory response and shift of half induction point under resource competition in genetic circuits. In 2021 IEEE 18th India Council International Conference (INDICON), pp. 1–6. IEEE (2021)
P. Chakraborty, M.K. Jolly, U. Roy, S. Ghosh, Spatio-temporal pattern formation due to host-circuit interplay in gene expression dynamics. Chaos Solitons Fractals 167, 112995 (2023)
X. Chen, Small RNAs and their roles in plant development. Ann. Rev. Cell Dev. 25, 21–44 (2009)
J.P. Crutchfield, K. Kaneko, Are attractors relevant to turbulence? Phys. Rev. Lett. 60(26), 2715 (1988)
J. Cursons, K.A. Pillman, K.G. Scheer, P.A. Gregory, M. Foroutan, S. Hediyeh-Zadeh, J. Toubia, E.J. Crampin, G.J. Goodall, C.P. Bracken et al., Combinatorial targeting by microRNAs co-ordinates post-transcriptional control of EMT. Cell Syst. 7(1), 77–91 (2018)
S. DiNardo, J. Heemskerk, S. Dougan, P.H. O’Farrell, The making of a maggot: patterning the drosophila embryonic epidermis. Curr. Opin. Genet. Dev. 4(4), 529–534 (1994)
Q. Dong, B. Hu, and C. Zhang. microRNAs and their roles in plant development. Front. Plant Sci. 13 (2022)
W. Driever, C. Nüsslein-Volhard, A gradient of bicoid protein in drosophila embryos. Cell 54(1), 83–93 (1988)
B. Ermentrout, Neural networks as spatio-temporal pattern-forming systems. Rep. Prog. Phys. 61(4), 353 (1998)
R.C. Friedman, K.K.-H. Farh, C.B. Burge, D.P. Bartel, Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19(1), 92–105 (2009)
R. Futahashi, H. Shirataki, T. Narita, K. Mita, H. Fujiwara, Comprehensive microarray-based analysis for stage-specific larval camouflage pattern-associated genes in the swallowtail butterfly, papilio xuthus. BMC Biol. 10(1), 1–22 (2012)
W. Ge, Q. Deng, T. Guo, X. Hong, J.-M. Kugler, X. Yang, S.M. Cohen, Regulation of pattern formation and gene amplification during drosophila oogenesis by the mir-318 microRNA. Genetics 200(1), 255–265 (2015)
A. Gierer, H. Meinhardt, A theory of biological pattern formation. Kybernetik 12, 30–39 (1972)
A. Gyorgy, J.I. Jiménez, J. Yazbek, H.-H. Huang, H. Chung, R. Weiss, D. Del Vecchio, Isocost lines describe the cellular economy of genetic circuits. Biophys. J. 109(3), 639–646 (2015)
H. Hamann, T. Schmickl, K. Crailsheim, Self-organized pattern formation in a swarm system as a transient phenomenon of non-linear dynamics. Math. Comput. Model. Dyn. Syst. 18(1), 39–50 (2012)
J. Hsia, W.J. Holtz, D.C. Huang, M. Arcak, M.M. Maharbiz, A feedback quenched oscillator produces turing patterning with one diffuser. PLoS Comput. Biol. 8(1), e1002331 (2012)
D. Jana, S. Batabyal, M. Lakshmanan, Self-diffusion-driven pattern formation in prey-predator system with complex habitat under fear effect. Eur. Phys. J. Plus 135(11), 1–42 (2020)
K. Jeyaseelan, K.Y. Lim, A. Armugam, Microrna expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke 39(3), 959–966 (2008)
M.W. Jones-Rhoades, D.P. Bartel, B. Bartel, Micrornas and their regulatory roles in plants. Annu. Rev. Plant Biol. 57, 19–53 (2006)
A. Koch, H. Meinhardt, Biological pattern formation: from basic mechanisms to complex structures. Rev. Mod. Phys. 66(4), 1481 (1994)
S. Kondo. How animals get their skin patterns: fish pigment pattern as a live turing wave. Syst. Biol. Chall. Complex. pp. 37–46 (2009)
S. Kondo, R. Asai, A reaction-diffusion wave on the skin of the marine angelfish pomacanthus. Nature 376, 765–768 (1995)
K.S. Kosik, The neuronal microRNA system. Nat. Rev. Neurosci. 7(12), 911–920 (2006)
N. Kumari, V. Kumar, Controlling chaos and pattern formation study in a tritrophic food chain model with cannibalistic intermediate predator. Eur. Phys. J. Plus 137(3), 1–23 (2022)
C.-J. Li, E.S. Liau, Y.-H. Lee, Y.-Z. Huang, Z. Liu, A. Willems, V. Garside, E. McGlinn, J.-A. Chen, T. Hong, Microrna governs bistable cell differentiation and lineage segregation via a noncanonical feedback. Mol. Syst. Biol. 17(4), e9945 (2021)
J. Li, G.-Q. Sun, Z.-G. Guo, Bifurcation analysis of an extended Klausmeier–Gray–Scott model with infiltration delay. Stud. Appl. Math. 148(4), 1519–1542 (2022)
Y. Liu, X. Tao, Z. Zhang, L. Zhu, A study of the turing pattern formation in a predator-prey model based on network and non-network environments. Eur. Phys. J. Plus 137(6), 691 (2022)
F.J. Lopes, F.M. Vieira, D.M. Holloway, P.M. Bisch, A.V. Spirov, Spatial bistability generates hunchback expression sharpness in the drosophila embryo. PLoS Comput. Biol. 4(9), e1000184 (2008)
B.T. Mbopda, S. Issa, S. Abdoulkary, R. Guiem, H. Fouda, Pattern formations in nonlinear dynamics of hepatitis B virus. Eur. Phys. J. Plus 136(5), 1–15 (2021)
J. Menezes, B. Moura, Pattern formation and coarsening dynamics in apparent competition models. Chaos Solitons Fractals 157, 111903 (2022)
H. Miyazako, Y. Hori, and S. Hara. Turing instability in reaction–diffusion systems with a single diffuser: characterization based on root locus. In 52nd IEEE Conference on Decision and Control, pp. 2671–2676. IEEE (2013)
H.G. Othmer, L. Scriven, Instability and dynamic pattern in cellular networks. J. Theor. Biol. 32(3), 507–537 (1971)
J.F. Palatnik, E. Allen, X. Wu, C. Schommer, R. Schwab, J.C. Carrington, D. Weigel, Control of leaf morphogenesis by microRNAs. Nature 425(6955), 257–263 (2003)
U. Roy, D. Singh, N. Vincent, C. K. Haritas, and M. K. Jolly. Spatiotemporal patterning enabled by gene regulatory networks. ACS Omega (2022)
P. Shu, C. Wu, X. Ruan, W. Liu, L. Hou, H. Fu, M. Wang, C. Liu, Y. Zeng, P. Chen et al., Opposing gradients of microRNA expression temporally pattern layer formation in the developing neocortex. Dev. Cell 49(5), 764–785 (2019)
G.-Q. Sun, H.-T. Zhang, Y.-L. Song, L. Li, Z. Jin, Dynamic analysis of a plant-water model with spatial diffusion. J. Diff. Equ. 329, 395–430 (2022)
X.-J. Tian, M. V. Ferro, and H. Goetz. Modeling ncRNA-mediated circuits in cell fate decision. Comput. Biol. Non-Coding RNA Methods Protocols, pp. 411–426 (2019)
X.-J. Tian, H. Zhang, J. Zhang, J. Xing, Reciprocal regulation between mRNA and microRNA enables a bistable switch that directs cell fate decisions. FEBS Lett. 590(19), 3443–3455 (2016)
A.M. Turing, The chemical basis of morphogenesis. Bull. Math. Biol. 52(1–2), 153–197 (1990)
Z. Zhang, Y.-W. Qin, G. Brewer, Q. Jing, Microrna degradation and turnover: regulating the regulators. Wiley Interdiscipl. Rev. RNA 3(4), 593–600 (2012)
Acknowledgements
PC and SG acknowledge the support by DST-INSPIRE, India, vide sanction Letter No. DST/INSPIRE/04/2017/002765 dated- 13.03.2019.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they do not have any known conflicts of interest.
Appendix
Appendix
1.1 List of parameters:
For better clarity, we have given a list of parameters used in our entire manuscript hereby in Tables 1, 2 and 3.
1.2 Model 2: Binary gene expression, temporal dynamics
To understand the temporal dynamics of the protein synthesized, we plot the time evolution curves of the protein starting from different initial conditions for three set of parameter values. For the parameter values of Fig. 14a, low synthesis state is the system’s stable steady state, starting from all initialization, the system converges to it. Similarly, 14c can be explained for its high synthesis stable state. However, the system has two steady states for the parameter value of 14b, and a bistable dynamics is shown in the output. Starting from different initial concentrations, the protein chooses any of its either low or high synthesis states, which one is more favorable and two drastic different concentrations coexist in output.
Time evolution curves of protein U for different initial conditions. x axis represents time and y axis represents the concentration of protein U. (a) The system is monostable with low synthesis state as a steady state. So for all initiation, protein U goes to its low synthesis stable state. (b) The system is bistable with two steady states. So with different initial conditions, the system chooses any of its nearest stable states and finally, we are left with two stable fixed states. (c) The system is monostable with its high synthesis stable state. All states from different initial conditions move to a single high synthesis stable state. Parameter \(\alpha\) has value 12 for (a), 13.3 for (b), 14 for (c). Rest of the parameter values for all (a)–(c) are \(\lambda =10,\; k=10,\;\delta =0.01,\;\phi =0.3\)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chakraborty, P., Ghosh, S. Quantitative modeling of diffusion-driven pattern formation in microRNA-regulated gene expression. Eur. Phys. J. Plus 138, 630 (2023). https://doi.org/10.1140/epjp/s13360-023-04258-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1140/epjp/s13360-023-04258-w