Abstract
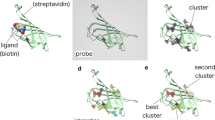
The paper deals with the identification of binding sites and concentrates on interactions involving small interfaces. In particular we focus our attention on two major interface types, namely protein-ligand and protein-peptide interfaces. As concerns protein-ligand binding site prediction, we classify the most interesting methods and approaches into four main categories: (a) shape-based methods, (b) alignment-based methods, (c) graph-theoretic approaches and (d) machine learning methods. Class (a) encompasses those methods which employ, in some way, geometric information about the protein surface. Methods falling into class (b) address the prediction problem as an alignment problem, i.e. finding protein-ligand atom pairs that occupy spatially equivalent positions. Graph theoretic approaches, class (c), are mainly based on the definition of a particular graph, known as the protein contact graph, and then apply some sophisticated methods from graph theory to discover subgraphs or score similarities for uncovering functional sites. The last class (d) contains those methods that are based on the learn-from-examples paradigm and that are able to take advantage of the large amount of data available on known protein-ligand pairs. As for protein-peptide interfaces, due to the often disordered nature of the regions involved in binding, shape similarity is no longer a determining factor. Then, in geometry-based methods, geometry is accounted for by providing the relative position of the atoms surrounding the peptide residues in known structures. Finally, also for protein-peptide interfaces, we present a classification of some successful machine learning methods. Indeed, they can be categorized in the way adopted to construct the learning examples. In particular, we envisage three main methods: distance functions, structure and potentials and structure alignment.
Similar content being viewed by others
References
J. Skolnick, M. Brylinski, Brief. Bioinform. 10, 378 (2009) DOI:10.1093/bib/bbp017.
G.R. Stockwell, J.M. Thornton, J. Mol. Biol. 356, 928 (2006).
B. Huang, Focus Struct. Biol. 8, 25 (2013).
M.L. Benson, R.D. Smith, N.A. Khazanov, B. Dimcheff, J. Beaver, P. Dresslar, J. Nerothin, H.A. Carlson, Nucl. Acids Res. 36, D674 (2008) DOI:10.1186/1471-2105-11-488.
J. Yang, A. Roy, Y. Zhang, Nucl. Acids Res. 41, D1096 (2012) DOI:10.1093/nar/gks966.
O.V. Kalinina, O. Wichmann, G. Apic, R.B. Russell, PLoS Comput. Biol. 7, e1002043 (2011).
I. Wallach, R. Lilien, Bioinformatics 25, 615 (2009) http://compbio.cs.toronto.edu/psmdb/.
G. Nicola, C.A. Smith, R. Abagyan, J. Comput. Biol. 15, 231 (2008).
Georgetown University Medical Center, Innovation center for biomedical informatics, http://icbi.georgetown.edu/biomedical/drug-discovery/ligand/.
J. Ito, Y. Tabei, K. Shimizu, K. Tsuda, K. Tomii, Nucl. Acids Res. 40, D541 (2012) http://possum.cbrc.jp/PoSSuM/database.html.
A. Loffet, J. Peptide Sci. 8, 1 (2002) DOI:10.1002/psc.366.
P. Vlieghe, V. Lisowski, J. Martinez, M. Khrestchatisky, Drug Discov. Today 15, 40 (2010) DOI:10.1016/j.drudis.2009.10.009.
V. Neduva, R. Linding, I. Su-Angrand, A. Stark, F. De Masi, T.J. Gibson, J. Lewis, L. Serrano, R.B. Russell, PLoS Biology 3, e405 (2005) DOI:10.1371/journal.pbio.0030405.
Rosetta Design Group, Macromolecular modeling blog, http://rosettadesigngroup.com/blog/742/the-structural-basis-of-peptide-protein-binding-strategies/.
D. Gfeller, FEBS Lett. 586, 2764 (2012).
F. Lampariello, G. Liuzzi, J. Optim. Theor. Appl. (2014) DOI:10.1007/s10957-014-0525-7.
I. Antes, Proteins: Struct. Funct. Bioinform. 78, 1084 (2010).
D. Duhovny, R. Nussinov, H. Wolfson, Efficient unbound docking of rigid molecules, in Proceedings of the 2nd Workshop on Algorithms in Bioinformatics (WABI) Rome, Italy, edited by Gusfield, Lecture Notes in Computer Science, Vol. 2452 (Springer Verlag, 2002) pp. 185–200.
D. Schneidman-Duhovny, Y. Inbar, R. Nussinov, H. Wolfson, Nucl. Acids Res. 33, 363 (2005).
P. Vanhee, J. Reumers, F. Stricher, L. Baeten, L. Serrano, J. Schymkowitz, F. Rousseau, Nucl. Acids Res. 38, D545 (2010) DOI:10.1093/nar/gkp893 http://pepx.switchlab.org/.
T. Gibson, F. Diella, H. Dinkel, K. Gould, C. Gemünd, C. Chica, S. Cameron, N. Blom, Phospho.ELM: a database of experimentally verified phosphorylation sites in eukaryotic proteins, http://phospho.elm.eu.org/.
T. Mi, J.C. Merlin, S. Deverasetty, M.R. Gryk, T.J. Bill, A.W. Brooks, L.Y. Lee, V. Rathnayake, C.A. Ross, D.P. Sargeant, C.L. Strong, P. Watts, S. Rajasekaran, M.R. Schiller, Nucl. Acids Res. 40, D252 (2012) DOI:10.1093/nar/gkr1189.
J.C. Obenauer, L.C. Cantley, M.B. Yaffe, Nucl. Acids Res. 31, 3635 (2003).
R. Amanchy, B. Periaswamy, S. Mathivanan, R. Reddy, S.G. Tattikota, A. Pandey, Nature Biotechnol. 25, 285 (2007).
T. Hertz, A. Bar-Hillel, D. Weinshall, Learning distance functions for image retrieval, in Proceedings of the 2004 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR04) (Washington DC, USA, 2004).
T. Shtatland, D. Guettler, M. Kossodo, M. Pivovarov, R. Weissleder, BMC Bioinform. 8, 280 (2007).
N. London, D. Movshovitz-Attias, O. Schueler-Furman, Structure 18, 188 (2010) DOI:10.1016/j.str.2009.11.012.
A. Shulman-Peleg, R. Nussinov, H.J. Wolfson, J. Mol. Biol. 339, 607 (2004).
J.A. Barker, J.M. Thornton, Bioinformatics 19, 1644 (2003) DOI:10.1093/bioinformatics/btg226.
T.A. Binkowski, Larisa Adamian, Jie Liang, J. Mol. Biol. 332, 505 (2003).
T.A. Binkowski, A. Joachimiak, J. Liang, Protein Sci. 14, 2972 (2005).
H. Yao, D.M. Kristensen, I. Mihalek, M.E. Sowa, C. Shaw, M. Kimmel, L. Kavraki, O. Lichtarge, J. Mol. Biol. 326, 255 (2003).
C. Hofbauer, A. Aszódi, J. Chem. Inf. Model. 45, 414 (2005) DOI:10.1021/ci0497049.
N. Kinoshita, J. Furui, H. Nakamura, J. Struct. Funct. Genomics 2, 9 (2001).
R. Minai, Y. Matsuo, H. Onuki, H. Hirota, Proteins Struct. Funct. Bioinform. 72, 367 (2008).
D. Kuhn, N. Weskamp, S. Schmitt, E.H. Hullermeier, G. Klebe, J. Mol. Biol. 359, 1023 (2006).
G. Ausiello, P.F. Gherardini, P. Marcatili, A. Tramontano, A. Via, M. Helmer-Citterich, BMC Bioinform. 9, S2 (2008) DOI:10.1186/1471-2105-9-S2-S2.
M. Jambonand, O. Andrieu, C. Combet, G. Deleage, F. Delfaud, C. Geourjon, Bioinformatics 21, 3929 (2005).
A.T.R. Laurie, R.M. Jackson, Curr. Protein Peptide Sci. 21, 1908 (2005).
S. Henrich, M.H. Outi Salo-Ahen, B. Huang, F.F. Rippmann, G. Cruciani, R.C. Wade, J. Mol. Recognit. 23, 209 (2010).
F. Xin, P. Radivojac, Curr. Protein Peptide Sci. 12, 456 (2011).
R. Nussinov, H.J. Wolfson, Proc. Natl. Acad. Sci. 88, 10495 (1991).
D. Fischer, H.J. Wolfson, S.L. Lin, R. Nussinov, Protein Sci. 3, 769 (1994).
A.C. Wallace, N. Borkakoti, J.M. Thornton, Protein Sci. 6, 2308 (1997).
J.S. Fetrow, J. Skolnick, J. Mol. Biol. 281, 949 (1998).
A.T.R. Laurie, R.M. Jackson, Bioinformatics 21, 1908 (2005).
M. Comin, F. Dellaert, C. Guerra, J. Comput. Biol. 16, 1577 (2009).
S.E. Leicester, J.L. Finney, R.P. Bywater, J. Math. Chem. 16, 315 (1994).
D.W. Ritchie, G.J.L. Kemp, J. Comput. Chem. 20, 383 (1999).
M.E. Bock, C. Garutti, C. Guerra, J. Comput. Biol. 14, 285 (2007).
W. Cai, X. Shao, B. Maigret, J. Mol. Graph. Modell. 20, 313 (2002).
R.J. Morris, R.J. Najmanovich, A. Kahraman, J.M. Thornton, Bioinformatics 21, 2347 (2005).
V. Cantoni, A. Gaggia, R. Gatti, L. Lombardi, Geometrical constraints for ligand positioning, in Proceedings of Bioinformatics - BIOSTEC (2011) pp. 26–29.
P.J. Besl, N.D. McKay, IEEE Trans. Pattern Anal. Mach. Intell. 14, 239 (1992).
A. Efrat, A. Itai, M.J. Katz, Algorithmica 31, 1 (2001).
M.L. Connolly, J. Appl. Crystallogr. 16, 548 (1983).
J. Liang, H. Edelsbrunner, C. Woodward, Protein Sci. 7, 1884 (1998).
P. Bertolazzi, C. Guerra, G. Liuzzi, BMC Bioinform. 11, 488 (2010) DOI:10.1186/1471-2105-11-488.
P. Brachetti, M. De Felice Ciccoli, G. Di Pillo, S. Lucidi, J. Global Optim. 10, 165 (1997).
L. Cirio, S. Lucidi, F. Parasiliti, M. Villani, J. Appl. Electromagn. Mech. 16, 13 (2002).
W.L. Price, A controlled random search procedure for global optimization, in Towards Global Optimization, Vol. 2, edited by L. Dixon, G. Szego (North-Holland, Amsterdam, 1978).
L. Ellingson, J. Zhang, PLoS ONE 7, e40540 (2012) DOI:10.1371/journal.pone.0040540.
P.J. Artymiuk, R.V. Spriggs, P. Willett, J. Am. Soc.r Inf. Sci. Technol. 56, 518 (2005).
C. Hofbauer, H. Lohninger, A. Aszodi, J. Chem. Inf. Comput. Sci. 44, 837 (2004).
R. Najmanovich, N. Kurbatova, J. Thornton, Bioinformatics 24, 105 (2008).
M. Shatsky, A. Shulman-Peleg, R. Nussinov, H. Wolfson, J. Comput. Biol. 13, 407 (2006).
N. Weskamp, E. Hllermeier, D. Kuhn, G. Klebe, ACM/IEEE Trans. Comput. Biol. Bioinform. 4, 310 (2007).
V. Vacic, L.M. Iakoucheva, S. Lonardi, P. Radivojac, J. Comput. Biol. 17, 55 (2010).
H. Deng, G. Chen, W. Yang, J.J. Yang, Proteins 64, 34 (2006).
N. Przulj, Bioinformatics 23, e177 (2007) DOI:10.1093/bioinformatics/btl301.
S.C. Bagley, R.B. Altman, Protein Sci. 4, 622 (1995).
A. Gutteridge, G.J. Bartlett, J.M. Thornton, J. Mol. Biol. 330, 719 (2003).
W.S. Valdar, Proteins 48, 227 (2002).
A.J. Bordner, Bioinformatics 24, 2865 (2008).
E. Petsalaki, A. Stark, E. García-Urdiales, R.B. Russell, PLoS Comput. Biol. 5, e1000335 (2009) DOI:10.1371/journal.pcbi.1000335.
M. Gribskov, A.D. McLachlan, D. Eisenberg, Proc. Natl. Acad. Sci. 84, 4355 (1987).
L.G. Trabuco, S. Lise, E. Petsalaki, R.B. Russell, Nucl. Acids Res. 40, W423 (2012) DOI:10.1093/nar/gks398.
D.J. Reiss, B. Schwikowski, Bioinformatics 20, i274 (2006).
J.S. Liu, A.F. Neuwald, C.E. Lawrence, J. Am. Stat. Assoc. 90, 1156 (1995).
T. Hertz, C. Yanover, BMC Bioinform. 7, S3 (2006) DOI:10.1186/1471-2105-7-S1-S3.
P.A. Reche, J.P. Glutting, H. Zhang, E.L. Reinher, Immunogenetics 56, 405 (2004).
L. Zhang, C. Shao, D. Zheng, Y. Gao, Mol. Cell. Proteomics 5, 1224 (2006).
S. Giguère, M.M., F. Laviolette, A. Drouin, J. Corbeil, BMC Bioinform. 14, 82 (2013) DOI:10.1186/1471-2105-14-82.
M. Nielsen, C. Lundegaard, T. Blicher, B. Peters, A. Sette, S. Justesen, S. Buus, O. Lund, PLoS Comput. Biol. 4, e1000107 (2008) http://dx.plos.org/10.1371.
P. Zhou, X. Chen, Y. Wu, Z. Shang, Amino Acids 38, 199 (2010) http://dx.doi.org/10.1007/s00726-008-0228-1.
Author information
Authors and Affiliations
Corresponding author
Additional information
Contribution to the Focus Point “Pattern Recognition Tools for Proteomics” edited by V. Cantoni.
Rights and permissions
About this article
Cite this article
Bertolazzi, P., Guerra, C. & Liuzzi, G. Predicting protein-ligand and protein-peptide interfaces. Eur. Phys. J. Plus 129, 132 (2014). https://doi.org/10.1140/epjp/i2014-14132-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1140/epjp/i2014-14132-1