Abstract.
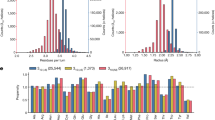
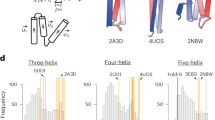
The stability of alpha helices is important in protein folding, bioinspired materials design, and controls many biological properties under physiological and disease conditions. Here we show that a naturally favored alpha helix length of 9 to 17 amino acids exists at which the propensity towards the formation of this secondary structure is maximized. We use a combination of thermodynamical analysis, well-tempered metadynamics molecular simulation and statistical analyses of experimental alpha helix length distributions and find that the favored alpha helix length is caused by a competition between alpha helix folding, unfolding into a random coil and formation of higher-order tertiary structures. The theoretical result is suggested to be used to explain the statistical distribution of the length of alpha helices observed in natural protein structures. Our study provides mechanistic insight into fundamental controlling parameters in alpha helix structure formation and potentially other biopolymers or synthetic materials. The result advances our fundamental understanding of size effects in the stability of protein structures and may enable the design of de novo alpha-helical protein materials.
Graphical abstract

Similar content being viewed by others
References
J.C. Kendrew, R.E. Dickerson, B.E. Strandberg, R.G. Hart, D.R. Davies, D.C. Phillips, V.C. Shore, Nature 185, 422 (1960)
T. Ackbarow, X. Chen, S. Keten, M.J. Buehler, Proc. Natl. Acad. Sci. U.S.A. 104, 16410 (2007)
L. Pauling, R.B. Corey, H.R. Branson, Proc. Natl. Acad. Sci. U.S.A. 37, 205 (1951)
Z. Qin, M.J. Buehler, Phys. Rev. Lett. 104, 198304 (2010)
Z. Qin, S. Cranford, T. Ackbarow, M.J. Buehler, Int. J. Appl. Mech. 1, 85 (2009)
M.J. Buehler, Y.C. Yung, Nat. Mater. 8, 175 (2009)
S.B. Prusiner, Science 216, 136 (1982)
M.A. DePristo, D.M. Weinreich, D.L. Hartl, Nat. Rev. Genet. 6, 678 (2005)
K.M. Pan, M. Baldwin, J. Nguyen, M. Gasset, A. Serban, D. Groth, I. Mehlhorn, Z.W. Huang, R.J. Fletterick, F.E. Cohen, S.B. Prusiner, Proc. Nat. Acad. Sci. U.S.A. 90, 10962 (1993)
S. Kumar, M. Bansal, Biophys. J. 75, 1935 (1998)
S. Penel, R.G. Morrison, R.J. Mortishire-Smith, A.J. Doig, J. Mol. Biol. 293, 1211 (1999)
W. Kabsch, C. Sander, Biopolymers 22, 2577 (1983)
H.M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T.N. Bhat, H. Weissig, I.N. Shindyalov, P.E. Bourne, Nucl. Acids Res. 28, 235 (2000)
A. Sali, E. Shakhnovich, M. Karplus, Nature 369, 248 (1994)
P.A. Thompson, W.A. Eaton, J. Hofrichter, Biochemistry 36, 9200 (1997)
V. Munoz, L. Serrano, Nat. Struct. Biol. 1, 399 (1994)
U.R. Doshi, V. Munoz, J. Phys. Chem. B 108, 8497 (2004)
V. Munoz, R. Ramanathan, Proc. Natl. Acad. Sci. U.S.A. 106, 1299 (2009)
S. Lifson, A. Roig, J. Chem. Phys. 34, 1963 (1961)
A. Vitalis, A. Caflisch, J. Chem. Theor. Comput. 8, 363 (2012)
J. Bertaud, J. Hester, D.D. Jimenez, M.J. Buehler, J. Phys.: Condens. Matter 22, 035102 (2010)
B.H. Zimm, J.K. Bragg, J. Chem. Phys. 31, 526 (1959)
E. Shakhnovich, Chem. Rev. 106, 1559 (2006)
K. Ghosh, K.A. Dill, J. Am. Chem. Soc. 131, 2306 (2009)
R. Burioni, D. Cassi, F. Cecconi, A. Vulpiani, Proteins 55, 529 (2004)
M. de Leeuw, S. Reuveni, J. Klafter, R. Granek, PLoS ONE 4, e7296 (2009)
D.U. Ferreiro, A.M. Walczak, E.A. Komives, P.G. Wolynes, Plos Comput. Biol. 4, e1000070 (2008)
M. Karplus, Y.Q. Zhou, D. Vitkup, J. Mol. Biol. 285, 1371 (1999)
L.D. Landau, E.M. Lifshitz, L.P. Pitaevskii, Statistical Physics, 3d revision and English edition (Pergamon Press, Oxford, New York, 1980)
T. Lazaridis, M. Karplus, Proteins-Struct. Funct. Genet. 35, 133 (1999)
M. Bertz, M. Wilmanns, M. Rief, Proc. Natl. Acad. Sci. U.S.A. 106, 13307 (2009)
C.-I. Brändén, J. Tooze, Introduction to Protein Structure, 2nd edition (Garland Pub., New York, 1999)
J.C. Phillips, R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, R.D. Skeel, L. Kale, K. Schulten, J. Comput. Chem. 26, 1781 (2005)
A. Barducci, G. Bussi, M. Parrinello, Phys. Rev. Lett. 100, 020603 (2008)
J.F. Marko, E.D. Siggia, Macromolecules 28, 8759 (1995)
J.R.C.v.d. Maarel, Introduction to Biopolymer Physics (World Scientific, Hackensack, NJ, 2008)
J. Bertaud, Z. Qin, M.J. Buehler, J. Strain Anal. Engin. Design 44, 517 (2009)
S. Keten, M.J. Buehler, Phys. Rev. Lett. 100, 198301 (2008)
M. Bonomi, D. Branduardi, G. Bussi, C. Camilloni, D. Provasi, P. Raiteri, D. Donadio, F. Marinelli, F. Pietrucci, R.A. Broglia, M. Parrinello, Comput. Phys. Commun. 180, 1961 (2009)
D.J. Barlow, J.M. Thornton, J. Mol. Biol. 201, 601 (1988)
S.M. Kreuzer, R. Elber, T.J. Moon, J. Phys. Chem. B 116, 8662 (2012)
P. Palencar, T. Bleha, Macromol. Theory Simul. 19, 488 (2010)
M.J. Buehler, Nature Nanotechnol. 5, 172 (2010)
P.R. LeDuc, W.C. Messner, J.P. Wikswo, Annu. Rev. Biomed. Engin. 13, 369 (2011)
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Qin, Z., Fabre, A. & Buehler, M.J. Structure and mechanism of maximum stability of isolated alpha-helical protein domains at a critical length scale. Eur. Phys. J. E 36, 53 (2013). https://doi.org/10.1140/epje/i2013-13053-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1140/epje/i2013-13053-8