Abstract
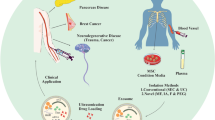
Exosomes, the subclass of small membrane extracellular vesicles, have great diagnostic and therapeutic potential, but the lack of standardized methods for their efficient isolation and analysis limits the introduction of exosomal technologies into clinical practice. This review discusses the problems associated with the isolation of exosomes from biological fluids, as well as the principles of traditional and alternative methods of isolation. The aim of the presented review is to illustrate the variety of approaches based on the physical and biochemical properties of exosomes that can be used for exosome isolation. The advantages and disadvantages of different methods are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Today, exosomes have become one of popular object for scientific research due to their unique natural properties that can be used for clinical purposes. Exosomes are a specific subclass of 50–100 nm extracellular vesicles (EV) naturally secreted by most eukaryotic cells. Endosomal biogenesis distinguishes them from other types of EV (apoptotic bodies and microvesicles) [1]. Exosomes are formed in endosomes by invagination of the endosomal membrane and then released into the extracellular space upon fusion of the formed multivesicular bodies with the cell plasma membrane [1].
These nanovesicles are surrounded by the lipid bilayer membrane enriched with etraspanins CD9, CD63, CD81, and CD83 that are traditionally used as exosomal markers [2]. Their internal contents include various cell-specific proteins, lipids, metabolites, and nucleic acids (DNA, mRNA, microRNA, long noncoding RNAs), partly reflecting the phenotype of exosome- producing cells. Exosomes mediate intercellular communication by transferring their contents to recipient cells and promoting various effects in these cells. Recipient cells can be located near or at a considerable distance from the place of secretion [3]. The delivery of exosome content from producing cells to recipient cells is targeted and loss-free [4]. It has been shown that exosomes are involved in a wide range of physiological and pathological processes: embryonic development, immune responses, tissue regeneration, vascular homeostasis regulation, and development of various diseases including cancer [5–8]. Since exosomes are found in many biological fluids of the body, the quantitative and qualitative analysis of circulating exosomes can be a promising tool for noninvasive diagnostic procedures, monitoring therapeutic efficacy, and preclinical relapse detection. The diagnostic potential of proteomic analysis and microRNA profiling of exosomes isolated from plasma, urine, saliva, and other fluids of patients with different diseases is being actively studied [9–12]. Another field of research is therapeutic potential of exosomes. The natural properties of exosomes such as small size, bloodstream stability, low immunogenicity, the ability to selectively transport their contents over long distances to recipient cells, as well as the possibility of modifying their surface and contents, determine the prospects of using these nanovesicles for targeted drug delivery [13–15], vaccine development [16, 17], and development of novel technologies in regenerative medicine [9, 18].
A key prerequisite for introducing exosomal technologies into clinical practice is the development of efficient and standardized isolation methods that allowing to obtain pure and homogeneous exosomal preparations in sufficient quantities. However, such methods are still lacking. The most widely used approaches today are all based on known physical (size, shape, density, charge) or chemical (composition of membrane surface) properties of vesicles. In recent years, a great number of studies have been performed with the aim of comparative analysis of different methods and their combinations for isolation of exosomes from various biological fluids [19–23]. These works have shown that exosomes isolated from the same biomaterial by different methods can vary significantly both in the yield and purity of particles and in the physical properties of nanovesicles (morphology, size) and in their biochemical composition (the level of surface markers, microRNA and proteins spectrum). On the other hand, the type of biological fluid also affects the result. For example, Langevin et al. compared the nanovesicles isolated by ultracentrifugation from the saliva, serum, and urine of healthy donors and showed that they differ in the non-coding RNAs expression profiles [21]. It is still unclear which isolation protocol may be optimal in each particular case, depending on the type of biological fluid and the final goal of research. Given the importance of the problem, work continues to develop new tools and protocols for the isolation of exosomes from various biological fluids.
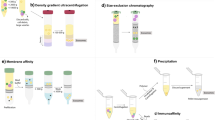
In the present review, we consider various methods for exosome isolation from biological fluids, both conventional (ultracentrifugation, filtration, gel chromatography, polymer precipitation, immunoaffinity separation) and relatively new: separation in a two-phase system, anion-exchange separation, oligo-phosphate aggregation (SubX-Matrix technology), precipitation with alginic acids, and silicon carbide solid-phase binding method. We also describe the principles of these methods, their advantages and disadvantages.
BASIC PROBLEMS OF EXOSOMAL ISOLATION FROM BIOLOGICAL FLUIDS
The high complexity of biological fluids and the lack of specific exosomal markers with high expression levels to differentiate these particles from other EV subtypes make the task of obtaining pure fractions of exosomes without loss much more difficult. Apart from exosomes, other components of similar size, mass and density circulate in any biological fluid: proteins, lipoproteins of different density, nucleoprotein complexes, other types of membrane vesicles, etc. These biologically active particles, as well as exosomes, can contain microRNAs or signal proteins and lipids. The exosomal pool itself is also quite heterogeneous [24]. It consists of subpopulations of nanovesicles that differ in size, morphology, surface markers, and biochemical contents. The contamination of exosomes with non-exosomal components of a biological fluid can lead to distortions in quantitative estimates of exosome contents [25]. On the other hand, some part of exosomes can be destroyed or lost during isolation, e.g., due to the aggregation of nanovesicles or the low level of expression of membrane markers, which will also skew the results.
The range of contaminating components can change depending on the source of exosomes. For example, lipoproteins and albumin, which are present in blood in significant amounts can be the major contaminants in case of isolation from blood. The concentration of lipoproteins in blood serum is considerably higher than that of circulating extracellular vesicles (1012 vs. 107–109 particles/mL) [26]. At the same time, it is incredibly difficult to separate exosomes from lipoproteins because of their similar sizes and densities. The main problem of isolation from urine is uromodulin (Tamm–Horsfall protein) as its concentration can reach 1.5 mg/mL. The polymer network formed by this protein can bind exosomes, reducing the efficiency of isolation [27].
EXOSOMAL ISOLATION METHODS
Ultracentrifugation
In experimental studies, the most commonly used method for isolation of exosomes is ultracentrifugation: separation of particle mixtures under the action of centrifugal force. The method is based on different sedimentation rates of particles that differ in size and density. There are three centrifugation-based protocols: differential centrifugation, rate-zonal gradient centrifugation, and isopycnic gradient centrifugation.
Differential ultracentrifugation (or differential velocity centrifugation) is the most optimal method for exosome isolation, which is currently considered the “gold standard” against which evaluate the effectiveness of other exosome isolation methodes. This technique usually involves several consecutive rounds of centrifugation with the increasing centrifugal force and centrifugation time, which allow separate particles of different sizes and densities. After each round, the supernatant is collected and subjected to further centrifugation. Although protocols may vary, most include four centrifugation steps: the first two steps (10 min at 300 g and then 10 min at 2000 g) precipitate cells, cell debris and large vesicles. The third step (30 min at 10 000 g) separates exosomes from non-exosomal vesicles, which are usually larger than 100–150 nm. Exosomes are precipitated in the last step by centrifugation at 100 000–150 000 g for 1–6 h. The advantages of this method include the possibility of exosome isolation from large volume of biomaterial, the relatively low cost, and the absence of additional chemical reagents for the procedure, which could contaminate exosome preparations. On the other hand, this method is not suitable for exosome extraction from small amounts of biomaterial. Disadvantages of the method are labor intensity, dependence of separation efficiency on the rotor type (fixed-angle or swinging-bucket) and its specific parameters (the maximum and minimum rotor radius, the length of the sedimentation path), temperature and viscosity of the initial liquid, which requires individual adjustment of standard centrifugation protocols depending on the rotor used and the properties of separated biofluids [28]. A significant disadvantage is also the presence of non-exosomal impurities in the exosome fraction. The method of differential centrifugation effectively separates only fractions of particles that differ significantly in sedimentation rates. Due to the high heterogeneity of composition of biological fluid (including exosome pool) containing components of the same density and size, some exosomes precipitate in the early stages of centrifugation together with larger particles, while other exosomes do not precipitate even after centrifugation at high g values, which leads to loss of some exosomes. Contrariwise, the non-exosomal components of biological fluid, such as lipoproteins, protein aggregates and other types of nanovesicles, can precipitate together with the exosome fraction. According to Kowal et al., 70% of the exosome fractions isolated by this method are 50–150 nm particles, 20% are larger than 150 nm, and 10% are smaller than 50 nm [29]. The combination of ultracentrifugation with additional purification steps (washing of the pellet with large volumes of buffer followed by repeated centrifugation, ultrafiltration of exosome suspension) can further purify the exosome fraction but at the cost of losing their number. The disadvantages of the method also include the possibility of exosomes damage during centrifugation that can change their morphology and functional properties.
More “pure” exosomes can be isolated by density gradient ultracentrifugation. This is a modified version of differential centrifugation. Unlike differential centrifugation, density gradient centrifugation separates of particles of similar size or density in a multicomponent sample. There are two types of density gradient ultracentrifugation: rate-zonal and isopycnic ultracentrifugation.
In rate-zonal centrifugation, particles are separated according to their sedimentation rate, which depends on particle size. In contrast to differential centrifugation, this method separates particles of different sizes simultaneously in single centrifugation step. The sample is loaded in a thin band on top of a buffer solution with a preformed concentration gradient gradually decreasing from the bottom of the tube to the meniscus and centrifuged. Under the action of centrifugal force, the particles move through the solution, as their density is higher than the solution density, and the speed of movement along the gradient depends on the size of the particle. As a result, the particles of similar sizes form discrete zones in the test tube. The density gradient increases separation efficiency by preventing premature sedimentation of particles, as well as zone mixing due convection currents. Centrifugation is performed until the optimal distribution of the zones in the test tube is achieved, and then the fractions are collected. Since the maximum density of the gradient buffer is lower than the density of particles in the sample, prolonged centrifugation may result in sedimentation of all components, including exosomes, on the bottom of the tube and, therefore, the time of centrifugation should be optimized.
Isopycnic gradient centrifugation is used to separate particles with different floatation densities and is based on the fact that particles in the medium with similar floatation densities remain stable. In this case, the sample is applied to the surface of the buffer concentration gradient overlapping the density range of all sample components. During centrifugation, particles move through the buffer gradient until they reach a position where their floatation density coincides with that of the buffer. The particles remain in this final position even after the rotor stops. Since the maximum density of the gradient is higher than the sample density, the particles do not reach the bottom even during prolonged centrifugation.
Exosomes are most often isolated in a sucrose gradient (2.0–0.25 M) and centrifuged at 210 000 g for 16 h. During centrifugation, all components including exosomes, apoptotic bodies, and protein aggregates move along the gradient until they reach a zone with a density corresponding to their floatation density. Exosomes are concentrated in the zone corresponding to their floatation density (1.10–1.18 g/mL), while protein aggregates and nucleoprotein complexes with the higher floatation density are concentrated closer to the bottom of the test tube [30]. The sucrose gradient makes it possible to increase the concentration of isolated exosomes more than threefold compared to conventional centrifugation [31]. The technique with a sucrose “cushion” formed of two sucrose layers of different concentrations, 1 and 2 M, has been proposed to purify exosomes from vesicles and complexes with similar floatation densities [32]. During centrifugation, the particles are distributed in layers depending on their size: large vesicles and aggregates are concentrated in the 2 M sucrose layer, exosomes are concentrated in the 1 M layer, and light molecules remain in the upper layers. According to some studies, the use of a 5–40% concentration gradient of iodixanol instead of sucrose can improve the purity of isolated exosomes [33]. Compared to sucrose, iodixanol (commercial name is OptiPrep) is more stable and less viscous [34]. In addition, the iodixanol gradient is isotonic at all concentrations used and, therefore, vesicles retain their shape and size when moving in the density gradient, which significantly increases separation efficiency [35]. Iodixanol density gradient centrifugation was shown to effectively separate exosomes from viral particles and apoptotic bodies [36]. This method allows effective separation of exosomes from similar-sized lipoproteins such as chylomicrons, very low, intermediate, and low-density lipoproteins because their floatation density is much lower (<1.063 g/mL). High-density lipoproteins are much smaller in size than exosomes but have a floatation density close to that of exosomes (1.06–1.21 g/mL); therefore, size-separation methods should be used to purify exosomes from these molecules [19], but this method is also rather low-productive and time-consuming, which limits its use in research.
Ultrafiltration
This technique is also size-based isolation methods. It involves on the procedure of filtering fluids through a filter with pores of a certain size. Exosome isolation protocols include several consecutive stages of filtration through filters with pore diameters of 0.8, 0.45, 0.22, sometimes 0.1 µm, resulting in the gradual removal of particles larger than the pore size from the filtered fluid. The most commonly used commercial filters are made of hydrophilic polymers with low affinity to proteins, such as the Millipore VVLP polyvinylidene fluoride filter with a pore size of 0.1 μm [37]. Ultrafiltration method makes it possible to obtain exosomes comparable to exosomes isolated by ultracentrifugation in such parameters as morphology and number of particles, contamination with non-exosomal proteins, and representation of exosome markers [38–40]. The advantage of the ultrafiltration technique is the simplicity of procedure, which does not require special equipment and skills, the high speed of purification that allows a large number of samples to be cleaned within a short period of time, and the absence of additional reagents that could contaminate exosomes The disadvantages of this technique are the possibility of exosome deformation and contamination with fluid components smaller than the pore size of the filter. In addition, some exosomal vesicles can be absorbed on the membrane, resulting in the loss of some exosomes, which is significant for isolation from small volumes of fluids [41]. The clogging of filter pores by proteins and other biopolymer molecules during filtration due to gradual concentration of contaminating molecules can reduce the the purification rate and lead to the loss of some part of exosomes. The ultrafiltration technology is used in commercial ExoMirTM Kits for the capture of exosomes (Bioo Scientific).
Gel Filtration Technique
The method of gel filtration, or exclusion chromatography, separates particles or molecules by the hydrodynamic radius due to their different abilities to penetrate gel pores of the stationary phase. This method has been effectively adapted to isolate exosomes from different types of liquids [42]. The method allows to separate small vesicles from large ones, as well as from unbound soluble proteins and high-density lipoproteins, because they are smaller than exosomes. Cross-linked agarose (Sepharose (CL-2B and CL-4B) and Sephacryl S-400) is most commonly used as a stationary phase. Compared to other methods, exosomes isolated by gel filtration are least contaminated with plasma proteins [43]. Moreover, as it has been shown in several studies, the degree of exosome purification from protein impurities depends on the used polymer and the column length [44, 45]. It should be noted that it is actually impossible to separate vesicles of similar sizes but different types, as well as to separate exosomes from other similar-sized biomolecules, by the method of gel filtration. Therefore, the exosomal fraction is often contaminated with small non-exosomal vesicles, large protein aggregates, and lipoproteins such as chylomicrons, low and very low density lipoproteins [19]. Another disadvantage of this method is a low yield of the target product, which is furthermore diluted. Nevertheless, despite some disadvantages related to contamination and losses, the gel filtration technique has significant advantages over other methods, especially in the context of their therapeutic and diagnostic applications and is considered by some authors as the most optimal isolation technique [42]. Since the particles during separation move in a liquid flow (commonly phosphate buffer, pH 7.4) only under the force of gravity, exosomes retain their integrity and functional activity [46]. No additional chemical reagents are used in the process of separation and, hence, no additional purification is required, as for example in case the isolating by PEG-precipitation.
Currently, there are several commercial exosome isolation kits based on gel filtration technique, e.g., SmartSECTM Single for EV Isolation (System Biosciences), qEV (Izon Science), PURE-EVs (Hansa Biomed). In oder to increase product yield and purity, Cell Guidance Systems has developed Exo-spinTM kit, combining PEG precipitation followed by purification by gel filtration.
Precipitation Techniques
The precipitation method is based on the aggregation of exosomes in the presence of precipitating agents. The procedure involves mixing a sample with a precipitant, followed by incubation and precipitation of resultant vesicle aggregates by low-speed centrifugation (1500 g); then the precipitate is dissolved in a buffer to a volume less than the initial one and used for further analysis.
PEG precipitation. Most popular precipitating agent is highly hydrophilic polymer polyethylene glycol (PEG). It has been used for a long time to precipitate viral particles, nucleic acids, and other biomolecules. The experimentally established phenomenon of precipitation in the presence of PEG is still poorly understood. The most popular theoretical models describing this process are the theory of excluded volume and the theory of attractive depletion forces [47]. According to the excluded volume model, molecules are precipitated as a result of a decrease in their hydration in the presence of the polymer [48]. The second model explains precipitation by the effect of attraction of molecules caused by the osmotic pressure of solution of polyethylene glycol [49]. The advantages of this method are its simplicity and rapidity of processing which allows simultaneous analysis of several samples, as well as minimal losses during extraction. Currently, PEG precipitation is second only to ultracentrifugation in terms of frequency of use. Usually, 8–12% of 6‑kDa PEG and 8–10% of 8-kDa PEG are used for exosome isolation [50, 51]. Unlike ultracentrifugation and ultrafiltration, during which vesicle shape can deform, the precipitation method allows obtaining exosomes of high morphological and functional quality. The main disadvantage of this technique is a low “purity” of exosomal preparation. This method only concentrates the exosomal fraction but does not separate it from other components of the sample. Exosome precipitate contains free proteins such as lipoproteins, immunoglobulins, as well as viral and other particles [52]. Polyethylene glycol impurities in the target product, as well as poor solubility of the precipitated aggregates, are also limitations for further analysis and application of the isolated exosomes.
The PEG precipitation technology is a basis for several commercial exosome isolation kits: Total Exosome Isolation Kit (Invitrogen), ExoQuick-TC Exosome Precipitation Solution (System Biosciences), miRCURY Exosome Kits (QIAGEN), Exo-Prep (HansaBioMed), PureExo Exosome Isolation kit (101Bio), ExoGAG (Nasasbiotech), Exosome Precipitation Solutions (Immunostep), miRCURY Exosome Isolation Kit (Exiqon). A significant disadvantage of commercial kits is their high cost, significant level of contamination of exosomes with nonexosomal components, and scaling limits. Meanwhile, some studies have shown that optimization of the standard PEG precipitation protocol (e.g., using PEG of different molecular weight and concentration, supplementing the protocol with additional washings and incubations) makes the procedure of exosome isolation even more efficient compared to commercial kits, though much cheaper [51–54]. In addition, this technique can be successfully used for isolation from large volumes of biofluid [50].
A variant of optimization the PEG precipitation technique that allows obtaining more “pure” exosome preparations is described in [55]. The authors suggested enriching the sample with membrane vesicles before adding PEG. For this purpose, the membranotropic/bifunctional agent Dextran Blue (2000 kDa) is added to the supernatant prepurified from cell debris by centrifugation. This agent initiates the aggregation of mostly membrane particles, while nonmembrane organoids, large biopolymers and supramolecular complexes remain in the solution. Next, PEG (20 kDa) is added and the exosomes are precipitated by low-speed centrifugation. Since the concentrations of Dextran Blue and PEG are rather low, the resultant exosomal preparations do not contain precipitating polymers that would impede further analysis.
MGP-precipitation (Mannuronate-Guluronate Polymer precipitation). An alternative technology for polymer-based precipitation of exosomes is described in [56]. In contrast to precipitation techniques with the formation of aggregates, this method is based on the “capture” of exosomes by the pores of alginate hydrogel formed of the homopolymer and heteropolymer residues of mannuronic and guluronic acids in the presence of calcium ions [57]. This method includes three main stages: mixing a polymer with a sample, short-term incubation of the mixture at room temperature, and exosome precipitation by low-speed centrifugation. Compared to PEG precipitation, exosomes isolated by MGP-precipitation are less contaminated with plasma proteins but more contaminated with large vesicles.
Charge-Based Precipitation
All extracellular vesicles are negatively charged under physiological conditions. This property of vesicles was a basis for the development of so-called “charge-based” (charge-dependent) precipitation techniques. It has been shown experimentally that exosome aggregation can be stimulated by the addition of positively charged protamine molecules [58]. Sodium acetate can cause the same effect [59]. The authors believe that sodium acetate breaks the hydration shell of exosomes and neutralizes their negative charge, which leads to vesicle aggregation by enhancing the hydrophobic interaction.
Affine Interactions
This group of methods is based on the ability of various biomolecules present on the surface of vesicles (lipids, polysaccharides, proteins) to interact affinely with other molecules, including antibodies, lectins and lipid-binding proteins.
Binding of membrane proteins. Various proteins, including tetraspanins CD9, CD63, CD81, CD82, heat shock proteins Hsp70 and Hsp90, major histocompatibility complex antigens (HLA-antigens), etc., are exposed on the surface of exosomal membranes. The antibodies against these proteins are used both for the analysis of the exosomal fraction and for the immunoprecipitation of exosomes. The antibodies used for immunoprecipitation are usually conjugated to a solid-phase carrier (latex, silicone), which can be in the form of polymer-coated magnetic particles (Dynabeads Magnetic Beads, Thermo Fisher Scientific), a centrifugation column (ExoTrap Exosome Isolation Spin Column, CosmoBio), modified pipette tips (MSIATMD.A.R.T.'S, Thermo Fisher Scientific), etc. The interaction between the antibody immobilized on the matrix surface and the exosomal membrane is relatively strong, allowing the bound exosomes to be washed from numerous components of biological fluid. The diversity of antibodies and carriers used in different formats explains the large number protocols based on affine interaction. For example, in the work of Clayton et al., the exosomes secreted by B-lymphocytes were isolated from the conditioned medium by magnetic separation using magnetic particles with conjugated HLA antibodies (DP, DQ, and DR) [60]. Magnetic particles were incubated with the conditioned medium for 24 h, then collected with a magnet and washed. The isolated particles were characterized by transmission electron microscopy and flow cytometry. 70% of the isolated vesicles were about 70 nm in size on the average, and 30% of them were larger than 100 nm. Another research team had concentrated vesicles from the conditioned medium of different cell lines using a hydrophilic polymer and the Total Exosome Isolation Reagent (Invitrogen) kit prior to magnetic separation of exosomes [61]. After 12-h incubation with the reagent and centrifugation, the precipitate was resuspended in phosphate buffered saline. The exosomes from the enriched preparation were isolated using CD9, CD63, and CD81 antibody-conjugated magnetic particles. Transmission microscopy, flow cytometry and Western blot analyses showed that the isolated preparation contained vesicles of the same or similar sizes with uniform morphology but no admixtures of proteins or protein aggregates.
In a number of works, an attempt has been made to automate the exosome isolation based on affine binding technology in order to speed up the process of their analysis. For example, Ueda et al. proposed a multichannel platform with MSIA D.A.R.T.'S Protein G tips (Thermo Fisher Scientific) to be used for this purpose [62]. These tips were originally designed for the automatic rapid isolation of antibodies followed by their mass spectrometric analysis. The tips contain a matrix of porous silica gel covalently bound to the recombinant G-protein which, in contrast to the native protein, has lost its ability to interact with albumin. This matrix does not bind albumin while being isolated from plasma, which increases the efficiency of detecting minor fractions of specific proteins. CD9 antibodies were additionally immobilized on silica gel for the specific “capture” of exosomes. The tip was filled with the serum (300 μL) and then the bound vesicles were washed and eluted from the matrix. Simultaneous isolation of vesicles from 12 samples takes only 30 min.
The main advantage of affine binding technique is the preparation purity due to specificity of antigen–antibody binding. This specificity is also a disadvantage as it promotes enrichment of the final product with exosomes with high levels of a certain surface marker and to the loss of exosomes with low levels of its expression. One more disadvantage of the method is a limitation associated with the availability of antibodies, as well as the difficulty of separating exosomes from the bound antibodies.
Aptamers. In affine binding, an alternative to antibodies can be aptamers: short artificial single-stranded oligonucleotides capable of highly specific recognition of particular target molecules due to formation of a unique 3D structure. The advantages of aptamers over antibodies are the possibility to select aptamers for almost any target and the possibility of their chemical modification, which makes it possible to obtain an aptamer with desired properties. Aptamers are cheaper than antibodies, have a long shelf life, and retain stability in a wide range of conditions.
Exosome isolation on the basis of affinity interaction between DNA aptamers and the surface exosomal marker CD63 is described in the works [63–65]. Zhang et al. used a biotin-modified CD63 aptamer immobilized on streptavidin-coated magnetic beads [63]. The resultant complex was incubated with plasma and then exosomes bound to the magnetic complex were collected with a magnet. To release the exosomes from the complex the authors added a nucleotide sequence complementary to CD63 aptamer. Hybridization of this sequence with CD63 aptamer leads to conformational disturbance of the latter, resulting in the release of exosomes. A similar approach was used in [64] to isolate exosomes from cell lines, except for the fact that the exosomes were separated from the magnetic complex by changing NaCl concentration. The possibility of isolating exosomes from urine with the CD63 aptamer was shown in [65]. An aptamer modified by thiol groups was immobilized on a titanium shell of magnetic particles (Fe3O4@TiO2-DNA aptamer) Double specific binding of the particles to the exosomes was achieved using both the aptamer and titanium dioxide TiO2, which was found to selectively bind to phosphate groups on the surface of lipid bilayer membranes of the exosomes. After the standard procedure of concentration on magnetic particles with a magnet, the exosomes were eluted by aptamer hydrolysis with DNAse and washing in 10% NH4OH (to break the titanium dioxide bond with phosphate groups). The authors have shown that the developed complex allows 92.6% of the exosomes to be isolated from urine in 10 min; they remain intact, contain the markers of urinary exosomes, and have a proteomic composition similar to that of exosomes isolated by ultracentrifugation.
Precipitation by Lectins
This technique is based on the specific and reversible binding of lectins (glycoproteins, mainly of plant origin) to carbohydrate residues (such as glycans) present on the surface of cells and membrane vesicles. The binding of lectins to membranes induces agglutination of vesicles. The resultant aggregates can be easily precipitated by low-speed centrifugation. Excessive simple sugars (glucose or mannose) are added to the precipitate to release individual exosomes from the aggregates. The sugars competitively break the bond between the polysaccharides of exosomes and lectins, and then the exosomes are washed from lectin. Lectins bind not only to exosomal membrane oligosaccharides and, therefore, all cells and large vesicles must be removed before adding lectin, e.g., by centrifugation or filtration. It should be noted that lectin-induced precipitation of exosomes is rarely used in research, and results are presented only in few methodological papers.
Lectins of different origin have been tested for precipitation of exosomes from different biological fluids, in particular, those isolated from cyanobacteria (OAA), potato (STL), wheat (WGA), and legumes (Con A) [66]. For example, the Solanum tuberosum (potato) lectin (STL) (Vector Laboratories) was used for exosome isolation from urine in the work by Royo et al. [67]. Urine samples were cleaned of cells and cell fragments by centrifugation at 2000 g and filtration through a membrane with a pore diameter of 0.22 μm. The pH value of purified samples was then adjusted to 7.5 and biotinylated lectin STL was added. After overnight incubation, the resultant STL/exosome complexes were collected using streptavidin-coated magnetic beads. The presence of exosomes in the preparations isolated by lectins was confirmed by cryo-electron microscopy and Western blot, but RNA levels in these preparations were much lower than in the preparations isolated using the Urine Exosome RNA Isolation Kit, Norgen Biotek (0.2 vs. 2.7 ng/mL urine), as well as ultracentrifugation and Total Exosome Isolation Reagent, Invitrogen (0.2 vs. 0.5 ng/mL urine). In general, the lectin precipitation technique is simple, does not require special equipment and expensive reagents, but is characterized by the low yield and low purity of the target product.
Precipitation by SubX-Matrix
This method also takes advantage of the properties of vesicular membrane, namely, the presence of phospholipid clusters on its surface. Both ends of a commercial SubX (Capital Biosciences) molecule can bind to a phosphate cluster. This property allows each SubX molecule to bind two adjacent exosomes through membrane phosphate residues, forming a vesicular dimer. Since membrane phospholipids contain many phosphate groups, the addition of excessive SubX to biological fluid causes the formation of micron-sized aggregates consisting of 10–15 exosomes, which are further efficiently precipitated by low-speed centrifugation [68]. Special buffer allows dissociation of precipitated aggregates back into the monomeric form and further analysis of exosomes. The possibilities of this technique for exosome isolation from plasma are presented in [69]. The comparative analysis of exosomes isolated by the SubX protocol and a number of conventional methods has shown that SubX makes it possible to obtain a relatively pure exosome fraction, being inferior only to the immunoprecipitation technique. However, the efficiency of this technique is relatively low: the yield of exosomes is more than 10 times lower compared to ultracentrifugation. At the same time, the technique is very simple and can be easily standardized and scaled-up for isolation from large volumes.
Separation in a Two-Phase Polyethylene Glycol–Dextran System
The method is based on selective distribution of biopolymers and their complexes between two phases formed in an aqueous solution by incompatible polymers such as, e.g., dextran and polyethylene glycol. It has been shown empirically that such a two-phase polymeric system makes it possible to concentrate vesicles mainly in the dextran phase and proteins, biopolymers and supramolecular complexes in the PEG phase [70]. For obtaining more pure exosome preparations, proteins can be re-extracted from the dextran phase by replacing the PEG phase. The ability of such two-phase system to selectively concentrate exosomes in the dextran phase has been demonstrated by Shin et al. [71, 72]. It should be noted that the distribution of biological particles or molecules in a two-phase system depends on many factors such as ionic strength, pH, the type, concentration and molecular weight of the polymers used, and the surface properties of particles; hence, on the one hand it is very difficult to predict system behavior in advance, but on the other hand such multifactorial nature makes the system extremely flexible, because systems with different separating properties can be obtained by manipulating the parameters. The possibility of optimizing the system for exosomal separation has been shown in [73].
Anion Exchange
Due to the negative surface charge under physiological conditions, exosomes can be electrostatically bound to a positively charged sorbent and then eluted from it with a buffer of high ionic strength. The evidence for the possibility of such an approach was obtained in [74]. Exosomes were isolated from the conditioned medium of different cell lines using a column with a monolithic polymeric material functionalized by quaternary ammonium groups (a strong anion-exchange sorbent). The authors have demonstrated that the method is not inferior to ultracentrifugation in the parameters such as the yield, quality and purity of the final product, but is much faster and simpler. In addition, it is much more effective in separating exosomes from proteins compared to ultrafiltration. It should be noted that this approach has yet been tested only in the conditioned media for cell cultures. Further studies are required to evaluate the potential of anion-exchange chromatography for the isolation of exosomes from more complex fluids such as plasma.
The alternative variant also based on the anion exchange platform was proposed in [75]. Exosomes from plasma were adsorbed on polycationic polymer (ExoCAS-2)-coated magnetic particles. The entire process of extraction takes 40 min and consists of three main stages. The plasma filtered through a 0.8-μm filter is mixed with the magnetic particles and incubated under shaking for 30 min. The exosomes bound to magnetic particles are collected with a magnet and washed from plasma proteins that were adsorbed together with the exosomes with a buffer, pH 6 (the experimentally selected value in the range of pI values of the protein). The exosomes were eluted from the particles with a high-salt solution (1 M NaCl). The analysis of isolated exosomes (size distribution, product yield, particle morphology, protein contamination level, microRNA composition) has shown that the ExoCAS-2 technique is superior to the methods such as ultracentrifugation and ExoQuick-TC Exosome Precipitation Solution (System Biosciences) and exoEasy (QIAGEN) commercial kits.
Solid-Phase Binding to Silicon Carbide
A fundamentally novel approach to exosome isolation was proposed by Norgen Biotek [76]. It is implemented in the following commercial kits: Plasma/Serum Exosome Purification Kit, Urine Exosome Purification Kit, Cell Culture Media Exosome Purification Kits, and Saliva Exosome Purification Kit. The method is based on the selective binding of proteins to negatively charged silicon carbide depending on the pH value of the solution and the value of the isoelectric point of the protein. The isoelectric point of exosomal surface proteins differs from the isoelectric points of histones and other proteins in biological fluids, which makes it possible, through optimization of pH, to selectively adsorb and then elute exosomes from the sorbent. The isolation procedure is very simple, fast, scalable, and requires no special equipment. It includes three stages: mixing with a sorbent (silicon carbide) in the binding buffer, precipitation of the sorbent with the bound exosomes by low-speed centrifugation, and elution of the exosomal fraction from the sorbent (isolation in a column is also possible). Isolated exosomes can be further used for the isolation of exosomal RNA and proteins. Extracellular vesicles isolated with SiC have considerably lower levels of contaminating materials such as macromolecular complexes, ribosomes and proteins, compared to extracellular vesicles isolated by ultracentrifugation or with precipitating agents. The comparative analysis of the Norgen Biotek technology and other commercial kits (ExoQuick-TC Exosome Precipitation Solution, System Biosciences and Total Exosome Isolation Reagent, Invitrogen), as well as ultracentrifugation, has shown that SiC significantly increases the yield of exosomal marker proteins and RNA, as well as the reproducibility of results, in case of isolation from urine [67].
CONCLUSIONS
In conclusion, various strategies are currently used for exosome isolation and purification, but there is no consensus on which of these methods is better. Meanwhile, the studies show that the final result strongly depends on the chosen isolation protocol, since each method has its own limitations affecting the purity and yield of exosomes. The choice of an appropriate protocol should depend on the goal of further research. For example, biomarker detection requires the techniques allowing isolation of “pure” exosomes from a small volume, while the functional integrity and number of vesicles are of paramount importance in the therapeutic situation. The ideal method of exosome isolation for clinical diagnostics has the following characteristics: low level of sample contamination, preservation of vesicle integrity, high yield, reproducibility, universality (isolation from all biological fluids), low cost, high rate of isolation simultaneously from a large number of samples (preferably for no more than one hour), accessibility and simplicity of equipment, and possibility of process automation [77]. All of the currently used standard methods of extraction are mainly time-consuming and labor-intensive, have an unstable yield of the target product, and require special sample preparation. However, the main problem is related to contamination of exosomes with various non-exosomal components of the biological fluids, especially in case of isolation from a complex fluid such as blood. The best option for solving the problem today seems to be combination of different isolation techniques; however, it considerably increases the time of analysis, limiting the use of such an approach for clinical purposes.
In recent years, several modern fractionation technologies have been adapted for exosome separation, e.g., Field Flow Fractionation (FFF), the method for fractionation of nanoparticles in a flow under the influence of fields of different physical nature (electric, magnetic, thermal, hydraulic, centrifugal, etc.). For example, Sitar et al. applied one of the FFF variants: Asymmetric Flow FFF (AF4), based on the separation of particles by size [78]. Exosomes isolated by this method retain their morphology and functionality, since fractionation occurs under mild conditions (without the stationary phase). This technique makes it possible not only to separate plasma exosomes from various macromolecules such as lipoproteins, proteins and other types of vesicles, but also to separate different subpopulations of exosomes [24, 79]. Another example of the relatively new technique for exosome isolation and purification is electrophoresis [80]. This is a charge-based separation of molecules. During electrophoresis, vesicles and their subpopulations can be fractionated on the basis of their electrophoretic mobility, i.e., taking into account not only their size but also their charge. Along with the development of techniques based on the new principles of particle separation, the possibilities of various microfluidic platforms using one or a combination of different separation principles and allowing simultaneous isolation and analysis of exosomes are now being actively studied [81].
A promising area of research in the field of exosomal technologies is a search for new universal biomarkers for these nanovesicles, in particular, membrane proteins [82]. Currently, tetraspanins CD9, CD63, and CD81 are widely used for exosome isolation. However, the expression level of these membrane proteins can vary greatly in different types of cells, which limits their application for immunoaffinity isolation of exosomes secreted by cell populations with the low expression levels of these proteins. The identification of new universal biomarkers that are abundant on the membranes of all exosomes regardless of their origin could be a basis for the development of a novel efficient exosome isolation technique.
REFERENCES
van Niel G., D’angelo G., Raposo G. 2018. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 19, 213–228.
Jeppesen D.K., Fenix A.M., Franklin J.L., Higginbotham J.N., Zhang Q., Zimmerman L.J, Liebler D.C., Ping J., Liu Q., Evans R., Fissell W.H., Patton J.G., Rome L.H., Burnette D.T., Coffey R.J. 2019. Reassessment of exosome composition. Cell. 177 (2), 428–445.
Tkach M., Théry C. 2016. Communication by extracellular vesicles: Where we are and where we need to go. Cell. 164, 1226–1232.
Mathieu M., Martin-Jaular L., Lavieu G., Théry C. 2019. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 21, 9–17.
Isola A., Chen S. 2016. Exosomes: The messengers of health and disease. Curr. Neuropharmacol. 15, 157–165.
Lin Y., Anderson J.D., Rahnama L.M.A., Gu S.V., Knowlton A.A. 2020. Exosomes and extracellular vesicles in cardiovascular physiology exosomes in disease and regeneration: Biological functions, diagnostics, and beneficial effects. Am. J. Physiol. Heart Circ. Physiol. 319 (6), H1162–H1180.
de Toro J., Herschlik L., Waldner C., Mongini C. 2015. Emerging roles of exosomes in normal and pathological conditions: New insights for diagnosis and therapeutic applications. Front. Immunol. 6, 203.
Dai J., Su Y., Zhong S., Cong L., Liu B., Yang J., Tao Y., He Z., Chen C. 2020. Exosomes: Key players in cancer and potential therapeutic strategy. Signal Transduct. Targeted Ther. 5, 145.
Popowski K., Lut, H., Hu S., George A., Dinh P.U., Cheng K. 2020. Exosome therapeutics for lung regenerative medicine. J. Extracel. Vesicles. 9 (1), 1785161.
Zarà M., Amadio P., Campodonic, J., Sandrini L., Barbieri S.S. 2020. Exosomes in cardiovascular diseases. Diagnostics. 10 (11), 943.
Yang H., Ma Q., Wang Y., Tang Z. 2020. Clinical application of exosomes and circulating microRNAs in the diagnosis of pregnancy complications and foetal abnormalities. J. Transl. Med. 18, 32.
Makler A., Asghar W. 2020. Exosomal biomarkers for cancer diagnosis and patient monitoring. Expert Rev. Mol. Diagn. 20 (4), 387–400.
Tran P. H., Wang T., Yin W., Tran T.T., Barua H.T., Zhang, Y., Midge S.B., Nguyen T.N.G., Lee B.-J., Duan W. 2019. Development of a nanoamorphous exosomal delivery system as an effective biological platform for improved encapsulation of hydrophobic drugs. Int. J. Pharm. 566, 697–707.
Liang Y., Duan L., Lu J., Xia J. 2021. Engineering exosomes for targeted drug delivery. Theranostics. 11, 3183–3195.
Salunkhe S., Dheeraj Basak M., Chitkara D., Mittal A. 2020. Surface functionalization of exosomes for target-specific delivery and in vivo imaging & tracking: Strategies and significance. J. Control. Release. 326, 599–614.
Xu Z., Zeng S., Gong Z., Yan Y. 2020. Exosome-based immunotherapy: A promising approach for cancer treatment. Mol. Cancer. 19, 160.
Barros F.M., Carneiro F., Machado J.C., Melo S.A. 2018. Exosomes and immune response in cancer: Friends or foes? Front. Immunol. 9, 730.
Maqsood M., Kang M., Wu X., Chen J., Teng L., Qiu L. 2020. Adult mesenchymal stem cells and their exosomes: Sources, characteristics, and application in regenerative medicine. Life Sci. 256, 118002.
Brennan K., Martin K., FitzGerald S.P., O’Sullivan J., Wu Y., Blanco A., Richardson C., Mc Gee M. 2020. A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Sci. Rep. 10, 1039.
Martins T.S., Catita J., Rosa I.M., da Cruz e Silva O.A.B., Henriques A.G. 2018. Exosome isolation from distinct biofluids using precipitation and column-based approaches. PLoS One. 13 (6), e0198820.
Langevin S.M., Kuhnell D., Biesiada J., Zhang X., Medvedovic M., Talaska G.G., Burns K.A., Kasper S. 2020. Comparability of the small RNA secretome across human biofluids concomitantly collected from healthy adults. PLoS One. 15 (4), e0229976.
Buschmann D., Kirchner B., Hermann S., Märte M., Wurmser C., Brandes F., Kotschote S., Bonin M., Steinlein O.K., Pfaffl M.W., Schelling. G., Reithmair M. 2018. Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing. J. Extracel. Vesicles. 7 (1), 1481321.
Mussack V., Wittmann G., Pfaffl M.W. 2019. Comparing small urinary extracellular vesicle purification methods with a view to RNA sequencing—Enabling robust and non-invasive biomarker research. Biomol. Detect. Quantif. 17, 100089.
Zhang H., Freitas D., Kim H.S., Fabijanic K., Li Z., Chen H., Mark M.T., Molina H., Martin A.B., Bojmar L., Fang J., Rampersaud S., Hoshino A., Irina Matei I., Kenific C.M., Nakajima M., Mutvei A.P., Sansone P., Buehring W., Wang H., Jimenez J.P., Cohen-Gould L., Paknejad N., Brendel M., Manova-Todorova K., Magalhães A., Ferreira J.A., Osório H., Silva A.M., Ashish Massey A., Cubillos-Ruiz J.R., Galletti G., Giannakakou P., Cuervo A.M., Blenis J., Schwartz R., Brady M.S., Peinado H., Bromberg J., Matsui H., Reis C.A., Lyden D. 2018. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 20, 332–343.
Sverdlov E.D. 2012. Amedeo Avogadro’s cry: What is 1 μg of exosomes? BioEssays. 34, 873–875.
Johnsen K.B., Gudbergsson J.M., Andresen T.L., Simonsen J.B. 2019. What is the blood concentration of extracellular vesicles? Implications for the use of extracellular vesicles as blood-borne biomarkers of cancer. Biochim. Biophys. Acta Rev. Cancer. 1871 (1), 109–116.
Fernández-Llama P., Khositseth S., Gonzales P.A., Star R.A., Pisitkun T., Knepper M.A. 2010. Tamm-Horsfall protein and urinary exosome isolation. Kidney Int. 77 (8), 736–742.
Livshits M.A., Khomyakova E., Evtushenko E.G., Lazarev V.N., Kulemin N.A., Semina S.E. 2015. Isolation of exosomes by differential centrifugation: Theoretical analysis of a commonly used protocol. Sci. Rep. 5, 17319.
Kowal J., Arras G., Colombo M., Jouve M., Morath J. P., Primdal-Bengtson B., Dingli F., Loew D., Tkach M., Théry C. 2016. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA. 113 (8), E968–E977.
Whiteside T.L. 2015. The potential of tumor-derived exosomes for noninvasive cancer monitoring. Expert Rev. Mol. Diagn. 15 (10), 1293–1310.
Singh K., Nalabotala R., Koo K.M., Bose S., Nayak R., Shiddiky M.J. 2021. Separation of distinct exosome subpopulations: Isolation and characterization approaches and their associated challenges. Analyst. 146, 3731–3749.
Raj D.A.A., Fiume I., Capasso G., Pocsfalvi G. 2012. A multiplex quantitative proteomics strategy for protein biomarker studies in urinary exosomes. Kidney Int. 81, 1263–1272.
Yu L.L., Zhu J., Liu J.X., Jiang F., Ni W.K., Qu L.S., Ni R.Z., Lu C.H, Xiao M.B. 2018. A comparison of traditional and novel methods for the separation of exosomes from human samples. BioMed. Res. Int. 2018, 3634563.
van Veldhoven P.P., Baumgart E., Mannaerts G.P. 1996. Iodixanol (optiprep), an improved density gradient medium for the iso-osmotic isolation of rat liver peroxisomes. Anal. Biochem. 237 (1), 17–23.
Li X., Donowitz M. 2008. Fractionation of subcellular membrane vesicles of epithelial and nonepithelial cells by OptiPrep™ density gradient ultracentrifugation. In: Exocytosis and endocytosis. Methods in Molecular Biology, 440. Eds. Ivanov A.I., New York: Humana Press, p. 97–110.
Cantin R., Diou J., Bélanger D., Tremblay A.M., Gilbert C. 2008. Discrimination between exosomes and HIV-1: Purification of both vesicles from cell-free supernatants. J. Immunol. Methods. 338 (1–2), 21–30.
Merchant M.L., Powell D.W., Wilkey D.W., Cummins T.D., Deegens J., Rood I.M., McAfee K.J., Fleischer C., Klein E., Klein J.B. 2010. Microfiltration isolation of human urinary exosomes for characterization by MS. Proteomics Clin. Appl. 4 (1), 84–96.
Gerlach J.Q., Krüger A., Gallogly S., Hanley S.A., Hogan M.C., Ward C.J., Joshi L., Griffin M.D. 2013. Surface glycosylation profiles of urine extracellular vesicles. PLoS One. 8 (9), e74801.
Alvarez M.L., Khosroheidari M., Kanchi Ravi R., Distefano J.K. 2012. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 82, 1024–1032.
Andreu Z., Rivas E., Sanguino-Pascual A., Lamana A., Marazuela M., González-Alvaro I., Sánchez-Madrid F., de la Fuente H. Yáñez-Mó M. 2016. Comparative analysis of EV isolation procedures for miRNAs detection in serum samples. J. Extracel. Vesicles. 5 (1), 31655.
Taylor D.D., Shah S. 2015. Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods. 87, 3–10.
Sidhom K., Obi P.O., Saleem A.A. 2020. A review of exosomal isolation methods: Is size exclusion chromatography the best option? Int. J. Mol. Sci. 21 (18), 6466.
Baranyai T., Herczeg K., Onódi Z., Voszka I., Módos K., Marton N., Nagy G., Mäger I., Wood M.J., Anda-loussi S.E.I., Pálinkás Z., Kumar V., Nagy P., Ágnes Kittel A., Buzás E.I., Ferdinandy P., Giricz Z. 2015. Isolation of exosomes from blood plasma: Qualitative and quantitative comparison of ultracentrifugation and size exclusion chromatography methods. PLoS One. 10 (12), e0145686.
Hagel L., Ostberg M., Andersson T. 1996. Apparent pore size distributions of chromatography media. J. Chromatogr. A. 743 (1), 33–42.
Arntz O.J., Pieters B.C., van Lent P., Koenders M.I., van der Kraan P.M., van de Loo F.A. 2020. An optimized method for plasma extracellular vesicles isolation to exclude the copresence of biological drugs and plasma proteins which impairs their biological characterization. PLoS One. 15 (7), e0236508.
Gámez-Valero A., Monguió-Tortajada M., Carreras-Planella L., Beyer K., Borràs F.E. 2016. Size-exclusion chromatography-based isolation minimally alters extracellular vesicles’ characteristics compared to precipitating agents. Sci. Rep. 6, 33641.
Lohmann L.J., Strube J. 2020. Accelerating biologics manufacturing by modeling: Process integration of precipitation in mAb downstream processing. Processes. 8 (1), 58.
Atha D.H., Ingham K.C. 1981. Mechanism of precipitation of proteins by polyethylene glycols. Analysis in terms of excluded volume. J. Biol. Chem. 256 (23), 12108–12117.
Marenduzzo D., Finan K., Cook P.R. 2006. The depletion attraction: An underappreciated force driving cellular organization. J. Cell Biol. 175 (5), 681–686.
Ludwig A.K., De Miroschedji K., Doeppner T.R., Börger V., Ruesing J., Rebmann V., Durst S., Jansen S., Bremer M., Behrmann E., Singer B.B., Jastrow H., Kuhlmann J.D., Magraoui F.E.I., Meyer H.E., Hermann D.M., Opalka B., Raunser S., Epple M., Horn P.A., Giebel B. 2018. Precipitation with polyethylene glycol followed by washing and pelleting by ultracentrifugation enriches extracellular vesicles from tissue culture supernatants in small and large scales. J. Extracel. Vesicles. 7 (1), 1528109.
Rider M.A., Hurwitz S.N., Meckes D.G. 2016. ExtraPEG: A polyethylene glycol-based method for enrichment of extracellular vesicles. Sci. Rep. 6, 23978.
Weng Y., Sui Z., Shan Y., Hu Y., Chen Y., Zhang L., Zhang Y. 2016. Effective isolation of exosomes with polyethylene glycol from cell culture supernatant for in-depth proteome profiling. Analyst. 141 (15), 4640–4646.
Garcia-Romero N., Madurga R., Rackov G., Palacin-Aliana I., Nunez-Torres R., Asensi-Puig A., Carrión-Navarro J., Esteban-Rubio S., Peinado H., González-Neira A., González-Rumayor V., Belda-Iniesta C., Ayuso-Sacido A. 2019. Polyethylene glycol improves current methods for circulating extracellular vesicle-derived DNA isolation. J. Transl. Med. 17 (1), 75.
Neerukonda S.N., Egan N.A., Patria J., Assakhi I., Tavlarides-Hontz P., Modla S., Muñoz E.R., Hudsond M.B., Parcells M.S. 2020. A comparison of exosome purification methods using serum of Marek’s disease virus (MDV)-vaccinated and-tumor-bearing chickens. Heliyon. 6 (12), e05669.
Konoshenko M.Y., Lekchnov E.A., Bryzgunova O.E., Kiseleva E., Pyshnaya I.A., Laktionov P.P. 2021. Isolation of extracellular vesicles from biological fluids via the aggregation–precipitation approach for downstream miRNAs detection. Diagnostics. 11 (3), 384.
Grunt M., Failla A.V., Stevic I., Hillebrand T., Schwarzenbach H. 2020. A novel assay for exosomal and cell-free miRNA isolation and quantification. RNA Biol. 17, 425–440.
Hay I.D., Rehman Z.U., Moradali M.F., Wang Y., Rehm B.H. 2013. Microbial alginate production, modification and its applications. Microb. Biotechnol. 6 (6), 637–650.
Deregibus M.C., Figliolini F., D’antico S., Manzini P.M., Pasquino C., De Lena M., Tetta C., Brizzi.M.F., Camussi, G. 2016. Charge-based precipitation of extracellular vesicles. Int. J. Mol. Med. 38 (5), 1359–1366.
Brownlee Z., Lynn K.D., Thorpe P.E, Schroit A.J. 2014. A novel “salting-out” procedure for the isolation of tumor-derived exosomes. J. Immunol. Methods. 407, 120–126.
Clayton A., Court J., Navabi H., Adams M., Mason M.D., Hobot J.A., Newman G.R., Jasani B. 2001. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J. Immunol. Methods. 247 (1–2), 163–174.
Oksvold M.P., Neurauter A., Pedersen K.W. 2015. Magnetic bead-based isolation of exosomes. In: RNA Interference. Methods in molecular biology (Methods and protocols). 1218. Eds. Sioud M. New York: Humana Press, p. 465–481.
Ueda K., Ishikawa N., Tatsuguchi A., Saichi N., Fujii R., Nakagawa H. 2014. Antibody-coupled monolithic silica microtips for high throughput molecular profiling of circulating exosomes. Sci. Rep. 4, 6232.
Zhang K., Yue Y., Wu S., Liu W., Shi J., Zhang Z. 2019. Rapid capture and nondestructive release of extracellular vesicles using aptamer-based magnetic isolation. ACS Sensors. 4 (5), 1245–1251.
Song Z., Mao J., Barrero R.A., Wang P., Zhang F., Wang T. 2020. Development of a CD63 aptamer for efficient cancer immunochemistry and immunoaffinity-based exosome isolation. Molecules. 25 (23), 5585.
Zhang N., Sun N., Deng C. 2021. Rapid isolation and proteome analysis of urinary exosome based on double interactions of Fe3O4@TiO2-DNA aptamer. Talanta. 221, 121571.
Liangsupree T., Multia E., Riekkola M.L. 2021. Modern isolation and separation techniques for extracellular vesicles. J. Chromatogr. A. 1636, 461773.
Royo F., Zuñiga-Garcia P., Sanchez-Mosquera P., Egia A., Perez A., Loizaga A., Arceo R., Lacasa I., Rabade, A., Edurne Arrieta E., Bilbao R., Unda M., Carracedo A., Falcon-Perez J.M. 2016. Different EV enrichment methods suitable for clinical settings yield different subpopulations of urinary extracellular vesicles from human samples. J. Extracel. Vesicles. 5 (1), 29497.
Malykh A.G., Malek A., Lokshin A., Evtushenko V. 2018. Simultaneous isolation of exosomes and cfDNA from liquid biopsies using universal kit based on SubX-Matrix TM technology. In: Proceedings of the American Association for Cancer Research Annual Meeting. 2018 Apr 14–18; Chicago, IL. Cancer Res. 2018. 78 (13 Suppl). Abstract nr 1618. https://doi.org/10.1158/1538-7445.am20181618
Shtam T., Evtushenko V., Samsonov R., Zabrodskaya Y., Kamyshinsky R., Zabegina L., Verlov N., Burdakov V., Garaeva L., Slyusarenko M., Nikiforova N., Konevega A., Malek A. 2020. Evaluation of immune and chemical precipitation methods for plasma exosome isolation. PLoS One, 15 (11), e0242732.
Kim J., Shin H., Kim J., Kim J., Park J. 2015. Isolation of high-purity extracellular vesicles by extracting proteins using aqueous two-phase system. PLoS One. 10 (6), e0129760.
Shin H., Park Y.H., Kim Y.G., Lee J.Y, Park J. 2018. Aqueous two-phase system to isolate extracellular vesicles from urine for prostate cancer diagnosis. PLoS One. 13 (3), e0194818.
Shin H., Han C., Labuz J.M., Kim J., Kim J., Cho S., Gho Y.S., Takayama S., Park J. 2015. High-yield isolation of extracellular vesicles using aqueous two-phase system. Sci. Rep. 5, 13103.
Slyusarenko M., Nikiforova N., Sidina E., Nazarova I., Egorov V., Garmay Y., Merdalimova A., Yevlampieva N., Gorin D., Malek A. 2021. Formation and evaluation of a two-phase polymer system in human plasma as a method for extracellular nanovesicle isolation. Polymers. 13 (3), 458.
Heath N., Grant L., De Oliveira T.M., Rowlinson R., Osteikoetxea X., Dekker N., Overman R. 2018. Rapid isolation and enrichment of extracellular vesicle preparations using anion exchange chromatography. Sci. Rep. 8, 5730.
Kim H., Shin, S. 2021. ExoCAS-2: Rapid and pure isolation of exosomes by anionic exchange using magnetic beads. Biomedicines. 9 (1), 28.
Haj-Ahmad Y., NorgenBiotek Corp. 2018. Methods for extracellular vesicle isolation and selective removal. United States Patent US10160964B2. 2018. Dec 25.
Abhange K., Makler A., Yi Wen Y., Ramnauth N., Mao W., Asghar W., Wan Y. 2021. Small extracellular vesicles in cancer. Bioact. Mater. 6 (11), 3705–3743.
Sitar S., Kejžar A., Pahovnik D., Kogej K., Tušek-Žnidarič M., Lenassi M., Žagar E. 2015. Size characterization and quantification of exosomes by asymmetrical-flow field-flow fractionation. Anal. Chem. 87 (18), 9225–9233.
Kim Y.B., Yang J.S., Lee G.B., Moon M.H. 2020. Evaluation of exosome separation from human serum by frit-inlet asymmetrical flow field-flow fractionation and multiangle light scattering. Anal. Chim. Acta. 1124, 137–145.
Morani M., Mai T.D., Krupova Z., Defrenaix P., Multia E., Riekkola M.L., Taverna M. 2020. Electrokinetic characterization of extracellular vesicles with capillary electrophoresis: A new tool for their identification and quantification. Anal. Chim. Acta. 1128, 42–51.
Lin B., Lei Y., Wang J., Zhu L., Wu Y., Zhang H., Wu L., Zhang P., Yang C. 2021. Microfluidic-based exosome analysis for liquid biopsy. Small Methods. 5 (3), 2001131.
Kugeratski F.G., Hodge K., Lilla S., McAndrews K.M., Zhou X., Hwang R.F., Zanivan S., Raghu Kalluri R. 2021. Quantitative proteomics identifies the core proteome of exosomes with syntenin-1 as the highest abundant protein and a putative universal biomarker. Nat. Cell Biol. 23 (6), 631–641.
Funding
The work was supported by the state task of the Ministry of Health of the Russian Federation “Development of a Technology and a Reagent Kit for Exosome Isolation from Human Biological Fluids Based on an Affine Phospholipid-binding Nanocomplex” (registration no. 121031100099-7).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by E. Makeeva
Rights and permissions
About this article
Cite this article
Yakubovich, E.I., Polischouk, A.G. & Evtushenko, V.I. Principles and Problems of Exosome Isolation from Biological Fluids. Biochem. Moscow Suppl. Ser. A 16, 115–126 (2022). https://doi.org/10.1134/S1990747822030096
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1990747822030096