Abstract
Structural studies of RNA–protein complexes are important for understanding many molecular mechanisms occurring in cells (e.g., regulation of protein synthesis and RNA-chaperone activity of proteins). Various objects investigated at the Institute of Protein Research of the Russian Academy of Sciences are considered. Based on the analysis of the structures of the complexes of the ribosomal protein L1 with specific regions on both mRNA and rRNA, the principles of regulation of the translation of the mRNA of its own operon are presented. The studies of the heterotrimeric translation initiation factor IF2 of archaea and eukaryotes are described, and the data on the interaction of glycyl-tRNA-synthetase with viral IRES are reported. The results of studying the interaction of RNA molecules with one of functionally important sites of the Hfq protein are presented, and the differences in the RNA-binding properties of the Hfq and archaeal Lsm proteins are revealed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
CONTENTS
Introduction
1. Regulatory Complexes of Ribosomal Protein L1 with mRNA
2. Determination of the Structure and Study of the RNA-Binding Properties of Global Translational Regulator Hfq
3. Study of the RNA-Binding Properties of Archaeal Lsm Proteins
4. Functional Study of the Transcriptional Regulators UxuR and ExuR
5. Structural and Functional Studies of Human Glycyl-tRNA-Synthetase
6. Structural Studies of the Heterotrimeric Translation Initiation Factor 2
Conclusions
INTRODUCTION
At the turn of the 20th and 21st centuries, the determination of the structures of bacterial ribosomes using X-ray diffraction (XRD) analysis and cryo-electron microscopy (Cryo-EM) became a breakthrough in the study of protein biosynthesis in cells. Determination of the structures of individual ribosomal components (specifically, ribosomal proteins and their complexes with fragments of ribosomal RNA) was an important starting point of this project. They made it possible to accelerate interpretation of the obtained immense electron density maps of the ribosomes and describe mobile parts of the ribosomal subunits, which cannot be observed in the structures of the whole ribosomes. Researchers from the Institute of Protein Research of the Russian Academy of Sciences (IPR RAS) have found the crystallization conditions for the 70S ribosomes and 30S ribosomal subunits from Thermus thermophilus and determined the structures of more than fifteen ribosomal proteins and several rRNA–protein complexes, which contributed greatly to the realization of the large-scale “ribosomal project”. A complex approach to determination of the structures of RNA–protein complexes was developed, which implies rational design of the RNA fragments forming most stable complexes with proteins; crystallization of these complexes; obtaining diffraction data on a Proteum (Bruker-AXS) laboratory system and on the BESSY, DESY, ESRF, and MAX IV synchrotron radiation sources; and solution of the structures using multiwavelength anomalous diffraction and multidomain molecular replacement method. The experience in the structural and functional studies of the rRNA–protein complexes made it possible to start solving new problems, concerning a considerable number of regulatory complexes. Study of the regulation of translation and transcription in cells at the atomic level makes it possible to answer new questions facing the scientific community. In this paper, we report briefly the results of the most interesting studies in this field of research that were performed at the Institute of Protein Research RAS.
1 REGULATORY COMPLEXES OF RIBOSOMAL PROTEIN L1 WITH mRNA
Ribosomal two-domain protein L1 is a part of the L1 protuberance of the large ribosomal subunit; in prokaryotes, this protein also controls the translation of the proteins of its operon according to the feedback principle. In E. coli, L1 protein binds the mRNA site before the first gene of the L11 operon [1]; in extremely thermophilic bacteria Thermus thermophilus and Thermotoga maritima, there are two binding sites on mRNA [2]; in archaea Methanococcus vannielii and Methanococcus jannaschii, the regulatory site on mRNA is located in the beginning of the coding sequence of the first gene of the L1 operon [3, 4]. The investigation of the regulatory properties of the ribosomal protein L1 became a natural continuation of the studies of the structural basis of protein biosynthesis.
L1 protein is the only ribosomal protein whose regulatory properties were investigated in detail using a comparative analysis of the structures of L1–rRNA and L1–mRNA complexes. Crystallized preparations of the L1–mRNA complex were obtained according to the procedure developed at the IPR RAS when studying the structure of the S8 ribosomal protein complex with a specific fragment of the 16S rRNA [5]. Ten mRNA fragments with lengths from 36 to 49 nucleotides, specifically bound with the L1 protein, were obtained. The length and nucleotide composition of two helices, flanking an asymmetric loop containing conservative nucleotides, were varied. The upper full-length helix–loop part of the mRNA fragments, which does not contain conservative nucleotides, was replaced in shortened versions by the four-nucleotide loop UUCG [6]. Seven fragments were variations of the L1-binding mRNA fragment of M. vannielii and three fragments were variations of the specific site of the mRNA of M. jannaschii. High mosaicity of crystals was decreased by adding mercury salts to the crystallization drop. The structures of three complexes were determined: the L1–mRNA complex (49 nt) from M. jannaschii (resolution 3.4 Å [7]) and two heterologic complexes, formed by the L1 protein from T. thermophilus and short mRNA fragments from M. vannielii with lengths of 38 (resolution 2.6 Å [8]) and 36 nt (resolution 2.1 Å [9]), respectively.
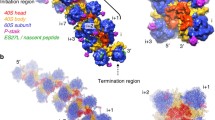
The L1 protein in the above complexes contacted mainly with the atoms of the sugar-phosphate mRNA backbone. A comparison of the structures of the ribosomal [10, 11] and obtained regulatory complexes of L1 protein showed that protein forms a network of inaccessible to solvent hydrogen bonds with a motif containing strictly conservative nucleotides and located at the structurally invariant to both mRNA and rRNA two RNA helixes junction. From the side of the L1 protein, adjacent loops of domain I are involved in this interaction (Fig. 1).
The specific features of interaction of L1 protein and specific mRNA and rRNA fragments were studied using a combination of structural analysis and surface plasmon resonance, which makes it possible to observe the molecular-interaction kinetics in real time [12]. The specific mRNA fragment is distinguished from the L1-binding rRNA fragment by strongly shortened sites, corresponding to the helix 78 and loop B of the ribosomal RNA (Fig. 2). This leads to reduction of affinity of the L1 protein to mRNA by several orders of magnitude in comparison with the L1–rRNA complex.
The main conservative contact region of L1 protein is located on its domain I and can be divided into two sites. One is the conservative triad Thr–Met–Gly, which forms inaccessible to solvent hydrogen bonds with nucleotides of the H77 rRNA helix [13]. The second site is the N-terminus of the α1 helix, which interacts with nucleotides of the H78 helix [14]. In the complexes of L1 protein with mRNA and rRNA, both interaction sites of domain I have identical conservative hydrogen bonds; however, the number of nonconservative contacts in the complex with mRNA decreases, reducing the protein affinity to the RNA matrix significantly. Domain I of L1 protein was obtained in an isolated form, and it was shown that it is sufficient for interaction with both rRNA and mRNA [15, 16] and has regulatory properties of the whole protein [17]. Amino acid residues of domain II of the protein in the ribosomal complex form additional contacts with rRNA, thus also increasing the stability of the ribosomal complex. This full-scale study of the complexes of L1 protein with mRNA and rRNA made it possible to explain the difference in the protein affinity to two different targets and reveal the basis of regulation of translation by the ribosomal protein L1 according to the feedback mechanism.
2 DETERMINATION OF THE STRUCTURE AND STUDY OF THE RNA-BINDING PROPERTIES OF GLOBAL TRANSLATION REGULATOR Hfq
The RNA-binding bacterial protein Hfq belongs to the abundant family of Sm/Lsm proteins and plays a role of a multifunctional translation-regulator protein in bacteria [18]. It is a small protein of ~10 kDa in size, which adopts a hexameric quaternary structure in solution [18]. In 1979, Hfq was identified as a host factor for replication of the RNA (+)-chain of bacteriophage Qβ [19]. Later, it was revealed that deletion of the hfq gene in Escherichia coli cells changes the production of more than 30 various proteins, including the DNA-reparation proteins [20], involved in iron metabolism [21] and synthesis of the σs subunit of bacterial RNA polymerase [22]. In most cases, translational regulation by Hfq is implemented by trans-coded small regulatory RNAs (sRNA). Hfq belongs to the so-called RNA chaperones, which include bacterial regulatory proteins CsrA, ProQ, KhpA, KhpB, and SpoVG, facilitating interaction of sRNAs with their targets on mRNA [23]. This regulation method makes it possible to control rapid adaptation of bacteria to environmental changes. Hfq is the most universal protein among RNA chaperones; it was found in most of bacteria. It stimulates the interaction of sRNA with complementary sites on mRNA, with subsequent repression or stimulation of translation, and controls the lifetime of mRNA due to the interaction with their 3'-poly(A) tails.
The first structures of Hfq from Staphylococcus aureus [24], shortened forms of the protein from E. coli [25], and full-length protein from Pseudomonas aeruginosa [26] were obtained in the beginning of the 2000s. Based on these results, several hypotheses about the facilitation of interaction between the two different RNAs by Hfq were proposed. Two main RNA-binding sites on the surface of the Hfq hexamer were identified: the uridine-specific site on the “proximal” protein surface (at the side of the N-termini of protein monomers) and the adenine-specific site on the opposite (“distal”) surface (Fig. 3).
Arrangement of the interaction sites of Hfq and RNA (the lateral view of the protein hexamer is at the center). The N- and C-termini of protein monomers are indicated. The following RNA binding regions are indicated: the uridine-specific proximal site, the adenine-specific distal site, and one out of six uridine-specific lateral sites. The following structures of the complexes of Hfq protein are shown: (top left) Hfq with the 5'-GUUUUUA-3' RNA [25] (view from the proximal site; PDB ID: 1KQ2), (bottom left) Hfq with the oligo(A) RNA [68] (view from the distal site; PDB ID: 3GIB), and (right) Hfq with UMP [28] (view from the proximal site; PDB ID: 4J6X). The RNA and nucleotide molecules are shown using a ball-and-stick model on the surface of Hfq protein hexamers.
The structures of the Hfq complexes with short RNA fragments made it possible to reveal the details of recognition of the RNA bases by amino acid residues of the protein and explain its high specificity to certain RNA sequences. It was interesting that the melting of the RNA secondary structure by Hfq did not require additional energy as ATP or GTP utilization; however, some biochemical data suggested ATPase activity of the protein [27]. We obtained the structure of the complex of Hfq with ATP and, taking into account the specific features of ATP contacts with amino acid residues of the protein, concluded that the probability of specific ATPase activity of Hfq is extremely low. Simultaneously, the ability of other single nucleotides to be bound with the protein surface was verified, and it was found that Hfq has high affinity to UTP and CTP; however, it does not bind GTP. The arrangement of bound nucleotides and orientation of their bases corresponded completely to those in the structures of Hfq complexes with oligonucleotides [28]. Based on these observations, the technique for mapping the bonding sites of single-stranded RNA on the surface of RNA-binding proteins was proposed [29, 30], which was used later when studying the homologous archaeal Lsm proteins. Moreover, the third (“lateral”) site of interaction of Hfq and RNA was identified (Fig. 3). It is located on the lateral surface of the protein hexamer at the N-terminal helix of the monomer and turns out to be the site for meeting and interaction of two RNAs: sRNA and mRNA. This finding was made simultaneously with the identification of this site by biochemical methods, which confirmed the reliability of our results [31–34]. Thus, we showed for the first time at the structural level how the fixation of RNA chain occurs, with its subsequent interaction with a protein partner. These data make a significant contribution to the further development of the model of interaction of sRNA and mRNA with participation of Hfq, and processes of translational regulation, mediated by the RNA-chaperone proteins.
3 STUDY OF THE RNA-BINDING PROPERTIES OF ARCHAEAL Lsm PROTEINS
As well as the bacterial Hfq proteins, the archaeal Lsm proteins (Sm Archaeal Proteins; SmAPs) belong to the Sm/Lsm family and have a similar tertiary structure, consisting of a five-stranded β-sheet with an N-terminal α-helix [35]. Genomes of various archaea can code from one to three SmAP paralogues, which are classified as SmAP1, SmAP2, and SmAP3. It is of interest that SmAP1 and SmAP2 from cells of the same species have a lower degree of homology than SmAP1 (or SmAP2) from cells of different organisms. However, they all have a rigorous consensus on the amino acid sequence RGXX (where X is a positively charged amino acid residue), in contrast to the YKHАI consensus of the bacterial Hfq (Fig. 4a). In addition, in contrast to most of Hfq, SmAP have elongated loop L4, which connects strands β3 and β4, and do not contain extended disordered C-terminus. The most significant difference between SmAPs and bacterial homologues is the formation of a quaternary structure in the form of homoheptamers (with few exceptions) rather than homohexamers (Fig. 4b). This property makes them close to their eukaryotic homologues: Sm/Lsm proteins, which are involved in RNA processing in cells and are generally part of various RNPs (e.g., splicesome). In contrast to bacterial and eukaryotic homologues, SmAP functions in archaeal cells have not been established. There are data indicating that they are bound several “immature” RNAs and some proteins, involved in RNA processing in cells [36–39], which suggests their possible engagement in RNA maturation; however, there are no direct proofs of the functional role of SmAP proteins.
(a) Alignment of the amino acid sequences of Hfq representatives (Hfq_Eco: E. coli, Hfq_Pae: P. aeruginosa, Hfq_Sty: Salmonella typhimurium, Hfq_Bsu: Bacillus subtilis), SmAP proteins (SmAP1_Mva: Methanococcus vannielii, SmAP1_Sso: Sulfolobus solfataricus, SmAP1-Afu: Archeoglobus fulgidus), and the eukaryotic Lsm protein from Saccharomyces cerevisiae. The elements of secondary structure corresponding to the amino acid sequences of proteins are shown at the bottom. The alignment was performed using the T-Coffee program, taking into account the data on the 3D structures of proteins (Espresso) [69]. (b) Schematic structures of (left) hexameric bacterial Hfq from P. aeruginosa PDB ID: 1U1S) and (right) heptameric archaeal SmAP from M. vannielii (PDB ID: 5MKI). View from the side of the α-helices of the protein monomers (proximal view).
The studies performed were aimed at determining the RNA-binding and RNA-chaperone properties of SmAPs in order to find possible functions of these proteins in archaea. After the determination of several structures of protein complexes with ribonucleotides, the RNA-binding sites on the surface of SmAPs were identified using the above-described technique. The results showed that SmAPs have only the uridine-specific RNA-binding site on the proximal side of the homoheptamers [40, 41]. Thus, one can hardly suggest that SmAPs can bind two different RNA molecules and participate in the translational regulation by sRNA. However, as we have shown, SmAPs can melt the structure of the target RNA without participation of the complementary RNA, which potentially allows these proteins to interact with mRNA, change its spatial structure, and thus affect the translation process. Therefore, one cannot exclude SmAPs from consideration as possible translational regulators in archaea; however, complete determination of their function requires additional studies.
4 FUNCTIONAL STUDY OF THE TRANSCRIPTIONAL REGULATORS UxuR AND ExuR
Gene expression is controlled at not only translational but also transcriptional level. An example is the regulation of the metabolic process of D-hexuronic acids assimilation, e.g., D-glucuronate and D-galacturonate (Ashwell metabolic pathway) [42, 43]. In bacteria, this pathway is an alternative to the most popular glycolytic metabolic pathway and plays a significant role for survival under stress conditions. The hexuronate utilization processes are controlled by two closely interrelated regulators: UxuR and ExuR [44, 45]. These proteins are paralogues, because ExuR apparently originated as a result of duplication of the uxuR gene [46], and belongs to the family of GntR regulators. To date, spatial structures of several proteins of this family have been obtained [46, 47]; however, the degree of their homology to UxuR and ExuR is no more than 25% [48]. The purpose of that study was to determine the structures of these two proteins and their complexes with regulatory DNA sites, in particular, in the presence of the ligands: glucuronate and galacturonate. Based on these structures, one can describe at the molecular level the regulation of the expression of genes involved in the metabolism of hexuronates.
The production of proteins in preparative amounts was hindered by the strong tendency of UxuR and ExuR to aggregation and their toxicity for producer cells; therefore, one had to choose carefully the conditions to produce the proteins [49]. The experiments aimed at determining the affinity of proteins to target DNA sites confirmed the presence of two different binding sites for UxuR and ExuR; the presence of sugars changed the protein affinity to DNA. The instability of proteins under crystallization conditions did not make it possible to obtain protein crystals in the free state and in complexes with ligands and/or DNA fragments; therefore, the structures of these complexes were modelled using flexible docking methods, based on the structures of homologous proteins [50]. As a result, a hypothetical site of binding sugars was localized in the region of the C-terminal UxuR domains, which coincided with the site predicted previously based on the mutagenesis data.
Using modern methods of molecular simulation and molecular dynamics, one can obtain interesting data on the molecular interaction; however, the reliability of the calculation models can be confirmed by only determination of the structures of complexes.
5 STRUCTURAL AND FUNCTIONAL STUDIES OF HUMAN GLYCYL-tRNA-SYNTHETASE
Enteroviruses and some related viruses initiate synthesis of their proteins in an infected eukaryotic cell according to cap-independent mechanism. In this case, the specific secondary structure of a certain site of viral mRNA (IRES) allows for assembly of a translational apparatus directly at a site close to the start codon. In contrast to the cap-dependent translation initiation, the mechanisms of IRES-dependent initiation are fairly diverse. IRESs from different sources can be absolutely different: neither a structural element common for all IRESs nor significant homology common for all their sequences have been found. This variety caused some difficulties in the IRES classification from the moment of their discovery. However, intense studies of viruses, their mRNA, and mechanisms for its translation have made it possible to reveal some common features to date, which allow for distinguishing quite definitely between IRES types.
Most viral IRESs have stable secondary and tertiary structures, facilitating their efficient binding with the 40S ribosomal subunit [51]. The translation initiation on all currently investigated IRESs of type I (IRES I) depends on the presence of special translational factors ITAF (IRES trans-acting factors), the list of which has not been completed yet. One of the ITAFs that are necessary for efficient initiation of the translation of polyovirus mRNA is human glycyl-tRNA-synthetase (GlyRS). This protein interacts with the apical part of the domain V of IRES polyovirus, which imitates the anticodon loop of the glycine tRNA and contains the glycine anticodon. GlyRS has a three-domain structure; an individual domain, referred to as ABD (Anticodon Binding Domain), which is responsible for the recognition of glycine anticodon. To date, the GlyRS structures both in the free form and in a complex with the glycine tRNA are known. This circumstance made it possible to study in detail the mechanism of recognition of the anticodon loop of tRNA by protein [52, 53]; however, the structure of the GlyRS complex with viral mRNA has not been determined yet.
Previously, the fragments domain V of the first-type IRES (IRES I) were obtained, and the minimum fragment interacting with GlyRS was determined [54]. It was also shown that human glycyl-tRNA-synthetase binds specifically fragments of IRES I viruses, which differ significantly in the taxonomic respect but have similar affinity. This fact indicates universality of the regulation mechanism of translation initiation using glycyl-tRNA-synthetase for these viruses and similarity of the spatial structures of the corresponding sites of their mRNAs [55].
The affinity of human glycyl-tRNA-synthetase to the fragments of IRES I exceeds that to the related tRNA by a factor of about 3 [55], a fact suggesting that the mechanisms of interaction between protein and RNA are similar in both cases. In contrast to tRNA, the anticodon loop at the apical part of the fifth IRES domain contains six (rather than five) nucleotides. The spatial structure of the domain V of the IRES I is still unknown. However, a theoretical model describing this fragment and its complex with the ABD domain of human glycyl-tRNA-synthetase (Fig. 5) has been suggested using existing structural data [56]. In comparison with the homologous part of tRNAGly the model implies a larger number of hydrogen bonds with protein, which explains the biochemical data obtained by us.
6 STRUCTURAL STUDIES OF THE HETEROTRIMERIC TRANSLATION INITIATION FACTOR 2
The translation initiation factor 2 (IF2) for bacteria is a six-domain monomeric protein, which facilitates binding of the initiator aminoacyl-tRNA with the small ribosome subunit. In archaea and eukaryotes, the translation initiation factor 2 (e/aIF2) consists of three different subunits: α, β, and γ. Functionally, it is similar to bacterial IF2; however, the e/aIF2 subunits do not exhibit homology with bacterial IF2. Subunits of eukaryotic eIF2 are larger than the homologous subunits of archaeal aIF2. In the presence of GTP, the heterotrimeric factor e/aIF2 recognizes specifically the initiation Met-tRNAi and forms a ternary complex e/aIF2–GTP–Met-tRNAi with it. In this form, e/aIF2 delivers Met-tRNAi to the P-site of small ribosome subunit, where exact recognition of the initiation codon on mRNA with subsequent GTP hydrolysis occurs due to the codon–anticodon interaction. In the complex with GTP, the initiation translation factor IF2 leaves ribosome, after which bound GDP is exchanged by GTP, and the factor recovers its ability to form a strong complex with Met-tRNAi. Thus, the translation initiation factor IF2 plays a critical role in the cells: it initiates protein synthesis on the ribosome.
The structure of heterotrimeric translation initiation factor IF2, which was determined for the first time at the IPR RAS [57], still remains the only crystal structure of full-length eukaryotic translation initiation factor eIF2. The aIF2 structure was determined from thermophilic archaea Sulfolobus solfataricus and used later to construct a model of the β-subunit of eukaryotic factor eIF2 in composition of the spatial structure of yeast pre-initiation complex 48S (48S PIC), which was investigated using cryoelectron microscopy [58]. The heterotrimeric translation initiation factor 2 is shaped as the letter L and consists of a conformationally-stable central part and two mobile wings [57]. The conformationally stable central part aIF2 (the γ-subunit with the third domain of the α-subunit aIF2αD3γ) is minimum and sufficient for binding by initiation Met-tRNAi.
Two crystal structures of the ternary complex aIF2–GTP–Met-tRNAi were deciphered almost simultaneously using archaeal translation initiation factor IF2. In 2012, a group of French researchers obtained crystals of the ternary complex, containing full-length heterotrimeric factor aIF2αβγ; however, they were not able to construct and refine the model for mobile β-subunit in view of unclear electron density [59]. In 2013, the crystals of the ternary complex, containing the core conformationally stable part of factor aIF2αD3γ, were obtained at the IPR RAS [60]. Both structures of the ternary complex made it possible to understand how aIF2 does distinguish between the initiation tRNA and elongation tRNA. The initiation tRNA is oriented with respect to the aIF2 protein by its D-side, whereas the elongation factor EF-Tu, which is structurally similar to aIF2γ, interacts with the elongation tRNA along the T-hairpin [61]. Thus, the differences in the nucleotide sequences of the T-hairpin of the initiation and elongation tRNAs may prevent binding of the initiation tRNA and EF-Tu, and differences in the D-arms may prevent binding of the elongation tRNA and aIF2.
A comparison of two structures of the ternary complex aIF2–GTP–Met-tRNAi showed that, despite the fact that tRNA is oriented with respect to protein by its D-side in both structures, it interacts with aIF2 in different ways (Fig. 6). In the structure of the ternary complex obtained by our group, the acceptor tRNA stem contacts with all three domains of the γ-subunit and aIF2αD3: CCA-end of the tRNA is localized in a gap between domains G(I) and III of the γ-subunit, domain II of the γ-subunit of aIF2 interacts with the 5′ end of Met-tRNAi, and the aIF2αD3 protein contacts with the tRNA elbow [60]. In the aIF2–GTP–Met-tRNAi model obtained in [59], the acceptor stem of the tRNA does not interact with the domain III of the γ-subunit, and the tRNA elbow contacts with aIF2αD1 and aIF2αD2, which (according to the biochemical data) possess nonspecific RNA-binding activity [62, 63].
The in vitro experiments performed in cooperation with the laboratory headed by I. Shatsky (Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University) showed that the archaeal factor aIF2 can maintain ribosomal scanning [64]. It was revealed using toeprinting that addition of archaeal factor aIF2 to the translation initiation system of mammals, in which its eukaryotic homologue is absent, leads to the formation of 48S initiation complex and mRNA scanning for the initiation AUG codon. This joint study made it possible to reveal the specific feature of the second stage of the translation initiation: attachment of a large ribosomal subunit to the 48S initiation complex. In this stage, additional factors are initiated (eIF5 and eIF5B), which promote attachment of a large subunit. In archaea, aIF5 is absent; however, we added eIF5 to the translation system with the archaeal second factor. It turned out that the 80S ribosome complex is not formed at replacement of eIF2 with aIF2. This result demonstrates that the stimulation of GTP activity of aIF2 in archaea differs from that in eukaryotes.
In our further studies devoted to the archaeal factor aIF2, a series of spatial structures of the γ-subunit of aIF2, representing different functional states of this ribosomal GTPase, were determined: from active (ON) GTP-bound state to inactive (OFF) GDP-bound state, passing through intermediate forms, where protein can be bound simultaneously with GDP and inorganic phosphate Pi or does not contain a nucleotide [65]. A comparison of the structures demonstrated for the first time displacement of domains II and III with respect to domain G(I) (although not so significant as in its structural homologue, translational elongation factor EF-Tu). Coordinated transformations occur also in the loops Switch 1 and Switch 2. The Switch 1 plays the role of a zipper for the nucleotide-binding pocket. It was found that conformation of this zipper does not make it possible to close completely the pocket in aIF2γ (in contrast to EF-Tu). It remains open during the entire cycle of aIF2γ operation and nothing hinders the γ-phosphate escape from protein after the GTP hydrolysis. This fact, in turn, indicates a possibility of spontaneous GTP hydrolysis for the factor aIF2 on the ribosome and explains the absence of factor aIF5, which stimulates the GTPase activity of aIF2 in archaea.
Three clusters of water molecules, forming water bridges between the γ-phosphate of GTP and two conservative residues Asp19 and Asp93, were found when analyzing the aIF2γ structure, whose nucleotide-binding pocket is in the intermediate state at the transition from the GTP-bound to GDP-bound form [66]. Those water bridges can be used to transfer the proton formed as a result of the catalytic reaction (GTP hydrolysis).
In addition, an extra loop (the third one) was revealed for the γ-subunit of aIF2, which can play the role of a switch during GTP hydrolysis [67]. As a result of a change in the conformation of this loop, the solvent takes an access to the γ-phosphate of GTP. The main gatekeeper on the path to the γ-phosphate of GTP is Met45. It is of interest that the eukaryotic γ-subunit of eIF2 contains a larger amino acid (isoleucine) instead of Met45, and additionally efforts are required to open the gate, which conditions the necessity of using the protein factor eIF5 inducing GTP hydrolysis. Note that the translation elongation factor EF-Tu does not contain this loop. All above data indicate that the GTPase-activating factor in archaea and spontaneous GTP hydrolysis for the aIF2 factor may be absent.
CONCLUSIONS
The experience of the researchers of the Institute of Protein Research of the Russian Academy of Sciences in structural and functional studies of RNA–protein complexes, accumulated within the “ribosomal project,” was successfully used in a series of studies of the principles of translation and transcription regulation of functionally important objects. This cycle includes the study of the autoregulation of translation of the universal-conservative ribosomal protein L1 in bacteria and archaea, the translational regulator Hfq and its archaeal homologues, the transcription regulators UxuR and ExuR; the investigation of the additional function of human glycyl-tRNA-synthetase; and the analysis of the heterotrimeric translation initiation factor IF2.
The results of the corresponding studies are briefly presented in the review. The nature of the different affinity of the ribosomal protein L1 to specifically recognizable mRNA and rRNA sites, which makes it possible to control the translation of its operon according to the feedback principle, was shown at the molecular level. For the Hfq protein, the structural basis of recognition of RNA molecules in the functionally important lateral RNA-binding protein site was revealed. The RNA-binding properties of a series of archaeal proteins of the Lsm family were investigated, and a significant difference in their RNA-binding properties in comparison with the bacterial homologue was shown. Structural data on the interaction of glycyl-tRNA-synthetase with viral IRES were obtained. Extensive studies of the heterotrimeric translation initiation factor IF2 in archaea and eukaryotes were performed.
Change history
19 April 2022
An Erratum to this paper has been published: https://doi.org/10.1134/S1063774521340095
REFERENCES
R. L. Gourse, H. A. de Boer, and M. Nomura, Cell 44 (1), 197 (1986). https://doi.org/10.1016/0092-8674(86)90498-8
A. O. Mikhailina, O. S. Kostareva, E. Yu. Nikonova, et al., Mol. Biol. 52 (1), 98 (2018). https://doi.org/10.7868/S0026898418010135
C. Mayer, C. Köhrer, P. Gröbner, and W. Piendl, Mol. Microbiol. 27 (2), 455 (1998). https://doi.org/10.1046/j.1365-2958.1998.00693.x
M. Hanner, C. Mayer, C. Köhrer, et al., J. Bacteriol. 176 (2), 409 (1994). https://doi.org/10.1128/jb.176.2.409-418.1994
S. V. Tishchenko, J. M. Vassilieva, O. B. Platonova, et al., Biochemistry 66 (9), 948 (2001). https://doi.org/10.1023/a:1012353122174
E. Ennifar, A. Nikulin, S. Tishchenko, et al., J. Mol. Biol. 304 (1), 35 (2000). https://doi.org/10.1006/jmbi.2000.4204
N. Nevskaya, S. Tishchenko, A. Gabdulkhakov, et al., Nucl. Acids Res. 33 (2), 478 (2005). https://doi.org/10.1093/nar/gki194
N. Nevskaya, S. Tishchenko, S. Volchkov, et al., J. Mol. Biol. 355 (4), 747 (2006). https://doi.org/10.1016/j.jmb.2005.10.084
S. Tishchenko, E. Nikonova, A. Nikulin, et al., Acta Crystallogr. D 62 (12), 1545 (2006). https://doi.org/10.1107/S0907444906041655
A. Nikulin, I. Eliseikina, S. Tishchenko, et al., Nat. Struct. Biol. 10 (2), 104 (2003). https://doi.org/10.1038/nsb886
S. Tishchenko, A. Gabdulkhakov, N. Nevskaya, et al., Acta Crystallogr. D 68 (8), 1051 (2012). https://doi.org/10.1107/S0907444912020136
O. Kostareva, S. Tishchenko, E. Nikonova, et al., J. Mol. Recognit. 24 (4), 524 (2011). https://doi.org/10.1002/jmr.1063
S. Tishchenko, O. Kostareva, A. Gabdulkhakov, et al., Acta Crystallogr. D 71 (2), 376 (2015). https://doi.org/10.1107/S1399004714026248
O. S. Kostareva, N. A. Nevskaya, S. V. Tishchenko, et al., Mol. Biol. 52 (1), 106 (2018). https://doi.org/10.7868/S0026898418010147
S. Tishchenko, E. Nikonova, V. Kljashtorny, et al., Nucl. Acids Res. 35 (21), 7389 (2007). https://doi.org/10.1093/nar/gkm898
S. Tishchenko, V. Kljashtorny, O. Kostareva, et al., J. Mol. Biol. 383 (2), 301 (2008). https://doi.org/10.1016/j.jmb.2008.08.058
A. P. Korepanov, O. S. Kostareva, M. V. Bazhenova, et al., Protein J. 34 (2), 103 (2015). https://doi.org/10.1007/s10930-015-9602-5
T. B. Updegrove, A. Zhang, and G. Storz, Curr. Opin. Microbiol. 30, 133 (2016). https://doi.org/10.1016/j.mib.2016.02.003
M. T. F. de Fernandes, W. S. Hayward, and T. J. August, J. Biol. Chem. 247 (3), 824 (1972).
D. Schuppli, G. Miranda, H. C. Tsui, et al., Proc. Natl. Acad. Sci. U. S. A. 94 (19), 10239 (1997). https://doi.org/10.1073/pnas.94.19.10239
B. Vecerek, I. Moll, T. Afonyushkin, et al., Mol. Microbiol. 50 (3), 897 (2003). https://doi.org/10.1046/j.1365-2958.2003.03727.x
A. Muffler, D. D. Traulsen, D. Fischer, et al., J. Bacteriol. 179 (1), 297 (1997). https://doi.org/10.1128/JB.179.1.297-300.1997
M. G. Jørgensen, J. S. Pettersen, and B. H. Kallipolitis, Biochim. Biophys. Acta Gene Regul. Mech. 1863 (5), 194504 (2020). https://doi.org/10.1016/j.bbagrm.2020.194504
M. A. Schumacher, R. F. Pearson, T. Møller, et al., EMBO J. 21 (13), 3546 (2002). https://doi.org/10.1093/emboj/cdf322
C. Sauter, J. Basquin, and D. Suck, Nucl. Acids Res. 31 (14), 4091 (2003). https://doi.org/10.1093/nar/gkg480
A. Nikulin, E. Stolboushkina, A. Perederina, et al., Acta Crystallogr. D 61 (2), 141 (2005). https://doi.org/10.1107/S0907444904030008
V. Arluison, S. K. Mutyam, C. Mura, et al., Protein Sci. 16 (9), 1830 (2007). https://doi.org/10.1110/ps.072883707
V. Murina, N. Lekontseva, and A. Nikulin, Acta Crystallogr. D 69 (8), 1504 (2013). https://doi.org/10.1107/S090744491301010X
M. Nemchinova, V. Balobanov, E. Nikonova, et al., Protein J. 36 (3), 157 (2017). https://doi.org/10.1007/s10930-017-9709-y
V. Balobanov, N. Lekontseva, A. Mikhaylina, and A. Nikulin, Methods Mol. Biol. 2113, 251 (2020). https://doi.org/10.1007/978-1-0716-0278-2_17
J. Chen, T. Morita, and S. Gottesman, Front. Cell. Infect. Microbiol. 9 (6), 201 (2019). https://doi.org/10.3389/fcimb.2019.00201
E. Sauer, S. Schmidt, and O. Weichenrieder, Proc. Natl. Acad. Sci. 109 (24), 9396 (2012). https://doi.org/10.1073/pnas.1202521109
K. E. Robinson, J. Orans, A. R. Kovach, et al., Nucl. Acids Res. 42 (4), 2376 (2013). https://doi.org/10.1093/nar/gkt1171
S. Panja, D. J. Schu, and S. Woodson, Nucl. Acids Res. 41 (15), 7536 (2013). https://doi.org/10.1093/nar/gkt521
V. N. Murina and A. D. Nikulin, Usp. Biol. Khim. 51, 133 (2011).
S. Fischer, J. Benz, B. Spath, et al., J. Biol. Chem. 285 (45), 34429 (2010). https://doi.org/10.1074/jbc.M110.118950
L. K. Maier, J. Benz, S. Fischer, et al., Biochim. 117, 129 (2015). https://doi.org/10.1016/j.biochi.2015.02.023
B. Märtens, G. Bezerra, M. Kreuter, et al., Life 5 (2), 1264 (2015). https://doi.org/10.3390/life5021264
B. Märtens, L. Hou, F. Amman, et al., Nucl. Acids Res. 45 (13), 7938 (2017). https://doi.org/10.1093/nar/gkx437
A. Nikulin, A. Mikhailina, N. Lekontseva, et al., J. Biomol. Struct. Dyn. 35 (8), 1615 (2017). https://doi.org/10.1080/07391102.2016.1189849
N. Lekontseva, A. Mikhailina, M. Fando, et al., Biochim. 175, 1 (2020). https://doi.org/10.1016/j.biochi.2020.05.001
G. Ashwell, Methods Enzymol. 5, 190 (1962). https://doi.org/10.1016/S0076-6879(62)05205-2
N. Peekhaus and T. Conway, J. Bacteriol. 180 (14), 3495 (1998). https://doi.org/10.1128/JB.180.14.3495-3502.1998
P. Ritzenthaler, C. Blanco, and M. Mata-Gilsinger, Mol. Gen. Genet. 199 (3), 507 (1985). https://doi.org/10.1007/BF00330766
C. B. Utz, A. B. Nguyen, D. J. Smalley, et al., J. Bacteriol. 186 (22), 7690 (2004). https://doi.org/10.1128/JB.186.22.7690-7696.2004
I. A. Suvorova, M. N. Tutukina, D. A. Ravcheev, et al., J. Bacteriol. 193 (15), 3956 (2011). https://doi.org/10.1128/JB.00277-11
S. B. Fillenberg, M. D. Friess, S. Korner, et al., PLoS 11 (6), e0157691 (2016). https://doi.org/10.1371/journal.pone.0157691
M. N. Tutukina, A. V. Potapova, P. K. Vlasov, et al., J. Biomol. Struct. Dyn. 34 (10), 2296 (2016). https://doi.org/10.1080/07391102.2015.1115779
T. A. Bessonova, N. Lekontseva, U. S. Shvyreva, et al., Protein Expr. Purif. 161, 70 (2019). https://doi.org/10.1016/j.pep.2019.05.001
Yu. A. Purtov, M. N. Tutukina, A. D. Nikulin, and O. N. Ozolin’, Biofizika 64 (1), 61 (2019). https://doi.org/10.1134/S0006350919010160
O. S. Nikonov, E. S. Chernykh, M. B. Garber, and E. Y. Nikonova, Biochemistry (Moscow) 82 (13), 1615 (2017). https://doi.org/10.1134/S0006297917130041
X. Qin, Z. Hao, Q. Tian, et al., J. Biol. Chem. 289 (29), 20359 (2014). https://doi.org/10.1074/jbc.M114.557249
X. Qin, X. Deng, L. Chen, and W. Xie, J. Mol. Biol. 428 (18), 3603 (2016). https://doi.org/10.1016/j.jmb.2016.05.018
E. Yu. Nikonova, A. O. Mikhailina, N. V. Lekontseva, et al., Biofizika 61 (2), 277 (2016). https://doi.org/10.1134/S0006350916020135
E. Yu. Nikonova, A. O. Mikhailina, M. S. Nemchinova, et al., Mol. Biol. 52 (1), 10 (2018). https://doi.org/10.7868/S0026898418010020
O. S. Nikonov, M. S. Nemchinova, V. G. Klyashtornyi, et al., Mol. Biol. 52 (1), 112 (2018). https://doi.org/10.7868/S0026898418010159
E. Stolboushkina, S. Nikonov, A. Nikulin, et al., J. Mol. Biol. 382 (3), 680 (2008). https://doi.org/10.1016/j.jmb.2008.07.039
J. L. Llácer, T. Hussain, L. Marler, et al., Mol. Cell. 59 (3), 399 (2015). https://doi.org/10.1016/j.molcel.2015.06.033
E. Schmitt, M. Panvert, C. Lazennec-Schurdevin, et al., Nat. Struct. Mol. Biol. 19 (4), 450 (2012). https://doi.org/10.1038/nsmb.2259
E. Stolboushkina, S. Nikonov, N. Zelinskaya, et al., J. Mol. Biol. 425 (6), 989 (2013). https://doi.org/10.1016/j.jmb.2012.12.023
P. Nissen, M. Kjeldgaard, S. Thirup, et al., Science 270 (5241), 1464 (1995). https://doi.org/10.1126/science.270.5241.1464
L. Yatime, E. Schmitt, S. Blanquet, and Y. Mechulam, J. Biol. Chem. 279 (16), 15984 (2004). https://doi.org/10.1074/jbc.M311561200
N. Pedulla, Nucl. Acids Res. 33 (6), 1804 (2005). https://doi.org/10.1093/nar/gki321
S. E. Dmitriev, E. A. Stolboushkina, I. M. Terenin, et al., J. Mol. Biol. 413 (1), 106 (2011). https://doi.org/10.1016/j.jmb.2011.08.026
O. Nikonov, E. Stolboushkina, V. Arkhipova, et al., Acta Crystallogr. D 70 (3), 658 (2014). https://doi.org/10.1107/S1399004713032240
O. Nikonov, O. Kravchenko, V. Arkhipova, et al., Biochimie 121, 197 (2016). https://doi.org/10.1016/j.biochi.2015.11.029
O. Nikonov, O. Kravchenko, N. Nevskaya, et al., Acta Crystallogr. D 75 (4), 392 (2019). https://doi.org/10.1107/S2059798319002304
T. M. Link, P. Valentin-Hansen, and R. G. Brennan, Proc. Natl. Acad. Sci. USA 106 (46), 19292 (2009). https://doi.org/10.1073/pnas.0908744106
C. Notredame, D. G. Higgins, and J. Heringa, J. Mol. Biol. 302 (1), 205 (2000). https://doi.org/10.1006/jmbi.2000.4042
ACKNOWLEDGMENTS
The authors are especially grateful to the pioneers of the structural and functional studies at the Institute of Protein Research of the Russian Academy of Sciences: M.B. Garber, S.V. Nikonov, and N.A. Nevskaya.
Funding
This study was supported by the Russian Foundation for Basic Research (RFBR) within the projects “Regulatory Complexes of Ribosomal Protein L1 with mRNA” (nos. 03-04-48327, 04-04-49634, 05-04-48338, and INTAS YSF no. 04-83-3842), “Study of the RNA-Binding Properties of the Hfq Protein” (nos. 04-04-48556, 07-04-00296, and 13-04-00783), “Study of the RNA-Binding Properties of the Archaeal Lsm Proteins” (no. 18-04-00222), “Structural and Functional Studies of Human Glycyl-tRNA-Synthetase” (no. 19-34-90135), and “Structural Studies of the Heterotrimeric Translation Initiation Factor IF2” (no. 18-04-01331) and the Russian Science Foundation within the projects “Functional Study of the Transcriptional Regulators UxuR and ExuR” (no. 18-14-00322), “Study of the RNA-Binding Properties of the Archaeal Lsm Proteins” (no. 14-14-00496), and “Structural and Functional Studies of Human Glycyl-tRNA-Synthetase” (no. 15-14-00028).
Author information
Authors and Affiliations
Corresponding author
Additional information
Translated by Yu. Sin’kov
The original online version of this article was revised due to a retrospective Open Access order.
Rights and permissions
Open Access. This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tishchenko, S.V., Mikhailina, A.O., Lekontseva, N.V. et al. Structural Investigations of RNA–Protein Complexes in Post-Ribosomal Era. Crystallogr. Rep. 66, 726–736 (2021). https://doi.org/10.1134/S1063774521050217
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1063774521050217