Abstract
Bacterial chaperonin GroEL is a complex ring-shaped protein oligomer that promotes the folding of other proteins by encapsulating them in the cavity. There is very little structural information about the disordered C-terminal fragment of the GroEL subunits, which is involved in the folding of the substrate protein. A 3D reconstruction of the GroEL apo-form was obtained by cryo-electron microscopy (cryo-EM) with a resolution of 3.02 Å and supplemented by molecular dynamics (MD) calculations. The results of cryo-EM and MD are in good agreement and demonstrate a diverse mobility of the protein subunit domains. The MD results predict the dynamics and the network of intramolecular contacts of the C-terminal sections of the protein. These results are of great importance for the subsequent study of the mechanism of protein folding in the GroEL cavity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Chaperones are proteins responsible for the folding of other macromolecules and for ensuring the formation of their native three-dimensional structure. Chaperones are also called Heat Shock Proteins (HSP), because their activity increases greatly with an increase in temperature [1]. The chaperone group includes proteins that have similar functions but differ in structure and substrate specificity [2]. One of the families of chaperones are chaperonins (HSP60). Usually they comprise two rings, each of which contains several subunits that differ in structure and their location in regard to different organisms [3].
Two groups of chaperonins are identified. The first group includes the chaperonins of bacteria, as well as the chaperonins of chloroplasts and mitochondria. They form symmetric rings, each of which consists of seven identical subunits. Chaperonins of the first group interact with a co-chaperone within their functional cycle, which forms a “lid” over the ring cavity to encapsulate the protein substrate. The second group of chaperonins combines proteins found in archaea and eukaryotic cytosol. Chaperonins of the second group also consist of two rings, but each of them includes eight or nine subunits. In addition, the subunits within one ring may differ in their structure [4]. Also, some bacteriophages have sequences in the genome that can encode GroEL orthologs [5, 6]. It is noteworthy that the bacteriophage chaperonins can significantly differ to bacterial ones: the chaperonin of bacteriophage OBP Pseudomonas fluorescens is an asymmetric single-ring structure consisting of seven subunits [7].
The most studied chaperonin is the prokaryotic GroEL/GroES complex, which, under normal conditions, is widely represented in E. coli cells. It interacts with non-native conformations of various proteins, preventing their improper folding and aggregation, and possesses weak ATPase activity [8]. Bacterial chaperonin GroEL is an intricate oligomeric protein complex consisting of 14 identical subunits with a molecular mass (MM) of 57 kDa each, combined into two rings of seven subunits. To properly perform its function, it needs to interact with GroES [9], which consists of seven identical subunits with MM 10 kDa each, connected into a domed ring structure. Each subunit of the GroEL heptameric ring comprises of three domains: apical, intermediate, and equatorial. The apical domain interacts with substrate proteins and GroES, the equatorial domain binds the ATP [10].
Contacts are formed between the amino acid residues of GroEL, some of which change during the functional cycle. In the apo-form (i.e., the form without nucleotides or a substrate protein), the rings are bound to each other by salt bridges between the residues of the equatorial domains: Lys105–Ala109 and Glu461–Arg452 [11–15]. Neighboring subunits within the (Arg197–Glu386) ring are also connected by salt bridges. It is believed that this contact, which stabilizes the structure of the oligomer in the apo-form, gets destroyed upon ATP-binding, which triggers conformational rearrangements of the protein complex [16, 17]. In addition, there are several intra-subunit salt bridges: Asp83–Lys327, Glu209–Arg58, and Glu409–Arg501. The first two bridges are most characteristic for chaperonins in the nucleotide-bound state in the absence of a co-chaperonin. Such structures are more open than the apo-form (the apical domain bends away from the equatorial one), but conformational rearrangements have not yet been completed. The contact between the Glu409 and Arg501 amino acids is very stable and does not break down even after ATP binding, or transition into the most open conformation of the subunit [16, 17].
Methods of X-ray crystallography and cryo-electron microscopy (cryo-EM) largely contributed to finding out the structure and mechanism of the GroEL/GroES complex, with the help of which more than 150 structures of the complex were obtained (according to the Protein Data Bank database), including structures captured at different stages of the functional cycle. However, structural methods have a number of limitations, in particular, they cannot decipher the mobile parts of molecules. In GroEL, 23 amino acids of the C-terminal region of each subunit form a mobile tail that emerges from the equatorial domain into the ring cavity [18]. This section is absent from the reconstructions of the complex, but is important for its functioning. The last 13 C-terminal amino acids consist of four repeating Gly-Gly-Met sequences and one methionine amino acid (hydrophobic sequence [GGM]4M), while the previous part of the tail contains negative charges. This hydrophobic sequence is conservative in all GroEL homologues [19]. The removal of this sequence does not cause the death of bacteria, but it reduces their resistance to heat shock [20]. Replacing the sequence with [AAA]4A led to the speed of substrate folding being lower than that of the wild type, but higher than of a mutant with completely removed C-termini [21]. The study of the hydrophilic part of the C-terminal fragment (amino acids 526–531), in which the hydrophilic section was replaced by a neutral or hydrophobic one, demonstrated that the mutants were significantly slower at restoring the native structure of the substrate protein rhodanase [22]. In other studies, it was shown that the rate of folding of Rubisco substrates [23] and GFP [24] significantly decreased when the C-ends were removed. Although structural methods did not provide results about the atomic structure of the C-terminal sites, they allowed us to detect some regularities in their dynamics. In [25], using cryo-EM, it was shown that the C-terminal fragments of the cis- and trans-rings deviate from each other and interact with the substrate dyes used in this experiment. In the same study, when examining the GroEL-Rubisco complex, it was noted that the substrate protein contacts with apical domains and C-termini.
Computational methods, including the method of molecular dynamics (MD), can complement structural methods by modeling flexible parts of the protein. Using MD, the C-terminal fragments of GroEL were previously studied and it was shown how their position changes during the functional cycle and how it is affected by the presence of nucleotides [26]. ATP- and ADP-bound crystal structures of GroEL were used in this study, and the positions of all of the residues, except for the C-terminal ones, were fixed. In this paper, the structure of the wild-type GroEL apo-form obtained by the authors with cryo-EM is used, an atomic model with C-terminal sections is constructed, and the MD system simulation is performed without additional restrictions. The MD results indicate differences in the mobility of the subunit domains, which is consistent with variations in the local resolution of the structure from cryo-EM, and also describe the dynamics of the C-terminal sites in the apo-form.
METHODS
Expression and purification of GroEL. The production of GroEL chaperonin was carried out using a frozen bacterial culture W3110 E. coli with a pOF 39 plasmid. Fifty mL of LB (Luria Broth, Sigma) bacterial medium containing 50 µg/mL of ampicillin was infected with a frozen culture and grown overnight at 37°С, stirred at 200 rpm. Next, six flasks of 200 mL of LB medium containing 50 µg/mL of ampicillin were taken and combined with a night culture to an optical density of 0.1 at 600 nm. GroEL production was carried out overnight at 37°С, stirred at 200 rpm. The resulting culture was centrifuged for 20 min at 5000g and 4°С. Cells were washed with 50 mM Tris-HCl buffer, pH 8.0, and resuspended in a lysing buffer (100 mM Tris-HCl, pH 8.1) containing 0.1 mM EDTA, 10 mM DTT, and 0.2 mg/mL of protease inhibitors (Sigma). Cells were destroyed by ultrasound treatment (Fisher Bioblock, Illkirch, France) with five 40 s impulses at an amplitude of 50%. The lysate was centrifuged for 30 min at 11 000 rpm, after which the supernatant was successively treated with dry (NH4)2SO4 up to 30 and 80%. Then, the specimen was centrifuged under the same conditions and the precipitate was dissolved in buffer B (50 mM Tris-HCl, pH 7.2, 0.1 mM EDTA, 2 mM DTT). The resulting solution was dialyzed against buffer B overnight. The dialysate was applied to a column with DEAE-Sepharose fast flow (Sigma), balanced with buffer B. Protein elution was carried out with a gradient of 0–500 mM NaCl. Chromatography was performed at a rate of 1–3 mL/min on the Akta Prime chromatographic system with Unicorn Control software (Amersham Biosciences, Piscatway, USA). Eight mL of fractions were collected and analyzed by electrophoresis in polyacrylamide gel in the presence of SDS. The resulting GroEL sample containing ∼350 mM NaCl (which stabilizes the protein) was heated in a round-bottomed flask in a water bath to 58°C, after which it was cooled to room temperature and Mg-ATP was added to the final concentration of 2 mM. Then, the mixture was heated to the same temperature for 2–3 min. This stage allows the GroEL to go through its natural cycle and release denatured E. coli proteins into the medium, which could be preserved in the chaperonin cavity. Denatured proteins were precipitated by centrifugation for 10 min at 11 000g. GroEL was dialyzed against buffer B overnight, and then re-purified on the DEAE-Sepharose fast flow according to the abovementioned method. Six mL fractions were collected and also analyzed by electrophoresis in polyacrylamide gel in the presence of SDS. Fractions containing purified GroEL were combined. To the purified GroEL substrate dry (NH4)2SO4 was added to 80% and stored at 4°C.
Obtaining the GroEL structure from cryo-EM data. The cryo-EM study was performed at the Resource Center for Probe and Electron Microscopy of the Kurchatov Institute Research Center. Initially, the supporting grids for electron microscopy with periodic holes in the amorphous carbon film (Quantifoil R1.2/1.3, Quantifoil) were subjected to a glow discharge in the PELCO easiGlow apparatus under standard conditions: sample processing time 30 s, current 0.25 mA, and residual pressure in the chamber 0.26 mBar. Next, 3 µL of the GroEL sample were applied to the grid and its vitrification was carried out in the Vitrobot Mark IV with the following parameters: compression force 0 units, Blot time from 2.5 to 3.5 s, temperature in the chamber 4.5°C, and humidity 95–100%. The frozen grids were then transferred to a Titan Krios 60-300 cryo-electron microscope equipped with a high-efficiency Falcon II electron detector, where images were captured using the EPU (FEI) software. Image processing and reconstruction were carried out using the Warp [27] and CryoSPARC [28] software packages. A total of 675 image stacks were taken, of which 590 stacks were selected for further analysis. Using the Warp software package, drift correction was performed, the parameters of the contrast transfer function (CTF) were evaluated, and also the selection of single projections of GroEL on the images took place. After pre-processing, 47 149 projections of single particles were isolated from the original images and exported to the CryoSPARC software package, where their two-dimensional classification was carried out. Next, 15 classes were selected; all particles were subjected to a 3D classification procedure. After that, 29 700 particles were selected, from which a 3D GroEL reconstruction was built after applying the D7 symmetry. The resolution of the structure estimated using FSC = 0.143 criterion was 3.02 Å.
Calculation of molecular dynamics trajectories. The atomic model of the GroEL apo-form for MD calculations is constructed in the COOT program [29] based on our cryo-EM structure. 23 residues of the C-terminal fragment (526–548), which were not detected in the density, were added manually using the standard Builder utility of the PyMOL program version 2.2.3 (www.pymol.org). The simulation was performed using the Gromacs software package [30] version 2020.1 in the a99SB-disp force field [31] at a constant temperature (300 K) and a constant pressure (1 atm), using periodic boundary conditions. The temperature and pressure were kept constant using V-rescale [32] and Parrinello-Rahman [33] algorithms, respectively. The protein was placed in a box with water (TIP4P-D model) of 17 × 17 × 18 nm containing 150 mM NaCl, including counterions to neutralize the total charge of the protein (519 Na+ and 253 Cl–). The electrostatic interactions were calculated using the PME algorithm with a cut-off radius of 1.2 nm. The cut-off radius for the Van der Waals forces was 1.2 nm. The integration time step was 2 fs. Before MD, system relaxation was performed, including an energy minimization stage, followed by heating the system from 5 to 300 K for 5 ns. The duration of the MD trajectory is 250 ns.
The values of RMSD and RMSF for Cα-atoms, as well as the pairwise distances between residues or atoms, are calculated using the standard utilities of the Gromacs package. As a marker for the existence of contact (as a “salt bridge”) between amino acids, a threshold of 0.3 nm was chosen, which corresponds to the sum of the Van der Waals radii sp3 N and sp3 O [34]. MDLovoFit software was used for differential calculation of RMSD [35]. Using MDLovoFit, structure fluctuations were analyzed by aligning all subsets of Cα-protein atoms corresponding to different fractions φ of the total number of Cα, and searching for the subgroup with the lowest RMSD value.
To compare the results of cryo-EM and molecular dynamics, a 3D map of the scattering density of chaperonin GroEL was constructed based on the MD trajectory. To do this, an algorithm was created for Python 3.6.9 [36], which translates a set of MD frames into a single 3D matrix. The dimensions of the matrix correspond to the abscissa, ordinate, and applicate of the space that contains the coordinates of the GroEL chaperonin, and each element of the matrix is proportional to the probability of finding atoms of the molecular model in the corresponding volume (voxel). In this work, the voxel size of the matrix corresponded to 1 × 1 × 1 Å, and only heavy atoms of the molecular model participated in the formation of the values of the matrix elements. For averaging, 25 000 frames of the molecular trajectory were used, which were pre-aligned by the progressive algorithm of the Gromacs software package. Twenty three amino acids located at the C-ends of the subunits were not taken into account during alignment.
RESULTS AND DISCUSSION
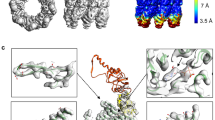
The average resolution of the GroEL tetradecamer structure obtained from cryo-EM data was 3.02 Å. Figure 1a shows the GroEL surface colored in accordance with the local resolution, which varies from 2.5 to 4.1 Å. Values from 2.5 to 3 Å are characteristic of the region of equatorial domains responsible for inter-ring interactions and ATP binding. In the zone of apical domains, which bind substrate proteins and GroES, the resolution drops to 4.1 Å. The distribution of the local resolution is consistent with the results of [37] and indicate a greater mobility of the apical domain relative to the equatorial one. Using the obtained scattering density map, an atomic model of the GroEL was constructed. Twenty three C-terminal amino acids were modeled and added to each subunit (Fig. 1b).
With this model, the MD trajectory with a length of 250 ns was calculated. Based on the trajectory, a 3D map of the scattering density of the GroEL chaperonin was constructed (Fig. 2), which, similar to the density from cryo-EM, reflects the probability of detecting an atom (scattering center) in the corresponding position of the structure. The scattering density map obtained from the MD trajectory is consistent with the density map from cryo-EM, demonstrating large values of matrix elements in the equatorial domain compared to the apical domain. The consent between the cryo-EM results obtained by averaging the structure over an ensemble of particles and the MD results obtained by averaging over time indicates that the number of particles collected by cryo-EM and the duration of the MD calculation are sufficient.
The calculation of the root-mean-square-fluctuations (RMSF) of the Сα atoms of each GroEL subunit in the MD trajectories showed that the domains differ in their mobility, and that the main contribution to conformational mobility comes from the amino acid residues of the C-terminal and the apical domain (Fig. 3). The average RMSF value for the equatorial domain is 0.15 nm, for the intermediate is 0.19 nm, for the apical is 0.27 nm, and for the C-terminal residues it exceeds 1 nm. According to the obtained data, the highest value of fluctuations of the ordered protein regions is observed for the residues of β-sheets 6 and 7 and α-helices K and L, located at the beginning and end of the apical domain, respectively, and can reach the value of 0.9 nm.
Differences in the mobility of GroEL domains during MD trajectories. The RMSF graph of the Cα-atoms of the protein subunit (a). The bold line shows the values averaged over 14 subunits. A pale wide band shows the range of RMSF values of each residue. The course of the main chain (only the Cα-atoms are shown) of the GroEL subunit (b), colored according to the scale of values of the B-factor (below).
The values of the root-mean-square deviations (RMSD) of the coordinates of the Сα-atoms from their position in the initial structure for all 14 subunits, according to the MD results, are in the range from 0.27 to 0.33 nm. Deviations from the initial position of the atoms of the apical domain significantly exceed those for the equatorial and intermediate domains: the average RMSD values are 0.27, 0.19, and 0.17 nm, respectively. To find out whether the calculated RMSD value of the apical domain is a result of an even greater deviation of its initial (starting) state from the equilibrium (achieved during MD) or of its greater mobility, an additional analysis of the RMSD atoms relative to their position in the previous structure was performed separately for each domain, as well as the alignment of various subsets of atoms of one subunit in order to identify the most variable regions using the MDLovoFit software package [35]. The analysis made it possible to identify a complete set of Сα -atoms of one subunit with the smallest RMSDs. It was found that at least 60% of the atoms of each subunit form a “fixed core” and can be superimposed on their original positions within 0.2 nm. As in [18], in this simulation, not all subunits underwent changes synchronously, which led to a changeable composition of the mobile and stationary parts. Nevertheless, for 11 of the 14 subunits, the set of fixed core residues coincides by 79%. Figure 4a shows the averaged change in time of an entire subunit (middle line) and the identified stable (lower line) and highly mobile sections (upper line). The atoms belonging to the mobile group deviate from the initial structure by more than 0.6 nm and are mainly part of the apical domain, as well as disordered C-ends (Fig. 4b, light). The fixed core quickly comes to an equilibrium position and does not change its structure during MD (Fig. 4b, dark).
Estimation of RMSD Cα-atoms of one subunit. RMSD as a function of the time of Cα-atoms: the entire subunits; 60% of the atoms that make up the fixed core of the subunits; the remaining 40% of the atoms undergoing the greatest changes. The corresponding pale color covers the ranges of RMSD values for 14 subunits (a). The superposition of MD frames. The stationary nucleus is shown as dark, 40% of the most mobile atoms are shown as light (b). The structures are oriented in a similar way to Fig. 3b. It can be seen that the mobile part includes the apical domain, C-ends and sections of loops of other domains. The distribution profile of the RMSD values of atoms relative to their position in the previous structure (Δt = 50 ps) of the equatorial, intermediate and apical domains (c).
RMSD analysis with respect to the preceding atomic location demonstrates a different rate of change in the position of atoms in different domains (Fig. 4c). During a selected interval of time (500 ps), the equatorial and intermediate domains change their structures within the range of 0.06–0.07 nm, while the changes in the RMSD of the apical domain lie in the range of 0.07–0.09 nm. Mobility analysis of the GroEL chaperonin based on MD results provides numerical estimates of the heterogeneous mobility of the domains of this protein and expands the qualitative picture of the structural dynamics of this macromolecule, obtained by cryo-EM.
Here we pay special attention to the dynamics of five physiologically significant amino acid contacts (Fig. 5). Despite the absence of direct contact between the side chains of the intraunit pairs of Arg58–Glu209 and Asp83–Lys327 residues in the starting model, these salt bridges are formed during the dynamics. The distance between a pair of heavy atoms of the Glu409–Arg501 salt bridge remains constant (0.28 nm) throughout the entire simulation. The lifetime of salt bridges formed between the subunits of one ring is ~95% of the MD time.
The stabilization of the intermolecular interaction of the rings is achieved due to two (in each pair of subunits) Arg452–Glu461 salt bridges. Analysis of the correlation between the formed contacts showed that the presence of one of the contacts in a pair is associated with the decrease in the probability of the formation of another (Table 1). The previously described Lys105–Ala109 interaction was not realized during MD, however, the Lys105–Ala109 residues of one subunit form a permanent hydrogen bond with the atoms of the main chain.
In the performed calculations, the highly mobile C-terminal fragments of the protein form an unordered network of contacts between themselves, and, also, interact with other domains of the subunits of their ring. The contact map (Fig. 6) shows that in addition to interacting with its subunit, the C-termini often bind to two neighboring subunits in the ring.
It has been noted that there is a difference in the frequency of contact formation between C-termini of one subunit with two neighboring subunits in the ring. It is more likely that the C-terminus binds to the inner surface of the next subunit located clockwise (when viewed from the apical domains). This observation can be explained by the protein structure, in which the C-terminus of the structured part of the equatorial domain of one subunit is located closer to the next clockwise subunit, which makes contact formation more likely. Note that the C-ends rarely form contacts with subunits located more than across one neighbor, although contacts between B and E subunits that are distant from each other were detected during the MD.
The study illustrates the effective interaction between experimental (cryo-EM) and theoretical (MD) approaches for studying the structural dynamics of a GroEL chaperonin as a model protein. The scattering density obtained by cryo-EM study is not only a starting point for creating a molecular model for conducting MD, but also allows us to qualitatively assess the relative mobility of individual domains of the protein complex. As shown in the paper, such a qualitative assessment can be compared with the result of MD calculations and be a way of verifying them. In turn, the MD method allows us to evaluate the quantitative characteristics of molecular mobility, the dynamic nature of the formation and breaking of specific bonds, as well as to consider extremely mobile parts of the protein, the structure of which cannot be reconstructed using cryo-EM.
Change history
19 April 2022
An Erratum to this paper has been published: https://doi.org/10.1134/S1063774521340058
REFERENCES
A. D. Maio, Shock 11 (1), 1 (1999). https://doi.org/10.1097/00024382-199901000-00001
D. Bose and A. Chakrabarti, IUBMB Life 69 (9), 647 (2017). https://doi.org/10.1002/iub.1656
L. Skjaerven, J. Cuellar, A. Martinez, and J. M. Valpuesta, FEBS Lett. 589, 2522 (2015). https://doi.org/10.1016/j.febslet.2015.06.019
H. Yebenes, P. Mesa, I. G. Munoz, et al., Trends Biochem. Sci. 36 (8), 424 (2011). https://doi.org/10.1016/j.tibs.2011.05.003
K. Hertveldt, R. Lavigne, E. Pleteneva, et al., J. Mol. Biol. 354 (3), 536 (2005). https://doi.org/10.1016/j.jmb.2005.08.075
L. P. Kurochkina, P. I. Semenyuk, V. N. Orlov, et al., J. Virol. 86 (18), 10103 (2012). https://doi.org/10.1128/jvi.00940-12
T. B. Stanishneva-Konovalova, P. I. Semenyuk, L. P. Kurochkina, et al., J. Struct. Biol. 209 (2), 107439 (2020). https://doi.org/10.1016/j.jsb.2019.107439
H. Sparrer, H. Lilie, and J. Buchner, J. Mol. Biol. 258 (1), 74 (1996). https://doi.org/10.1006/jmbi.1996.0235
H. Grallert and J. Buchner, J. Struct. Biol. 135 (2), 95 (2001). https://doi.org/10.1006/jsbi.2001.4387
A. L. Horwich, G. W. Farr, and W. A. Fenton, Chem. Rev. 106 (5), 1917 (2006). https://doi.org/10.1021/cr040435v
G. H. Lorimer, X. Fei, and X. Ye, Philos. Trans. R. Soc. London B 373 (1749), 20170179 (2018). https://doi.org/10.1098/rstb.2017.0179
S. J. Ludtke, M. L. Baker, D.-H. Chen, et al., Structure 16 (3), 441 (2008). https://doi.org/10.1016/j.str.2008.02.007
N. A. Ranson, D. K. Clare, G. W. Farr, et al., Nat. Struct. Mol. Biol. 13 (2), 147 (2006). https://doi.org/10.1038/nsmb1046
H. R. Saibil, W. A. Fenton, D. K. Clare, and A. L. Horwich, J. Mol. Biol. 425 (9), 1476 (2013). https://doi.org/10.1016/j.jmb.2012.11.028
B. Sot, A. Galán, J. M. Valpuesta, et al., J. Biol. Chem. 277 (37), 34024 (2002). https://doi.org/10.1074/jbc.m205733200
J. Ma, P. B. Sigler, Z. Xu, and M. Karplus, J. Mol. Biol. 302 (2), 303 (2000). https://doi.org/10.1006/jmbi.2000.4014
N. A. Ranson, G. W. Farr, A. M. Roseman, et al., Cell 107 (7), 869 (2001). https://doi.org/10.1016/s0092-8674(01)00617-1
S. Piana and D. E. Shaw, J. Phys. Chem. B 122 (49), 11440 (2018). https://doi.org/10.1021/acs.jpcb.8b07366
L. Brocchieri and S. Karlin, Protein Sci. 9 (3), 476 (2008). https://doi.org/10.1110/ps.9.3.476
N. F. Mclennan, A. S. Girshovich, N. M. Lissin, et al., Mol. Microbiol. 7 (1), 49 (1993). https://doi.org/10.1111/j.1365-2958.1993.tb01096.x
Y.-C. Tang, H.-C. Chang, A. Roeben, et al., Cell 125 (5), 903 (2006). https://doi.org/10.1016/j.cell.2006.04.027
K. Machida, A. Kono-Okada, K. Hongo, et al., J. Biol. Chem. 283 (11), 6886 (2008). https://doi.org/10.1074/jbc.m708002200
J. Weaver and H. S. Rye, J. Biol. Chem. 289 (33), 23219 (2014). https://doi.org/10.1074/jbc.m114.577205
S. Ishino, Y. Kawata, H. Taguchi, et al., J. Biol. Chem. 290 (24), 15042 (2015). https://doi.org/10.1074/jbc.m114.633636
D.-H. Chen, D. Madan, J. Weaver, et al., Cell 153 (6), 1354 (2013). https://doi.org/10.1016/j.cell.2013.04.052
K. M. Dalton, J. Frydman, and V. S. Pande, PLoS One 10 (3), e0117724 (2015). https://doi.org/10.1371/journal.pone.0117724
D. Tegunov and P. Cramer, Nat. Methods 16 (11), 1146 (2019). https://doi.org/10.1038/s41592-019-0580-y
A. Punjani, J. L. Rubinstein, D. J. Fleet, and M. A. Brubaker, Nat. Methods 14 (3), 290 (2017). https://doi.org/10.1038/nmeth.4169
P. Emsley, B. Lohkamp, W. G. Scott, and K. Cowtan, Acta Crystallogr. D 66 (4), 486 (2010). https://doi.org/10.1107/s0907444910007493
M. J. Abraham, T. Murtola, R. Schulz, et al., SoftwareX 1, 19 (2015). https://doi.org/10.1016/j.softx.2015.06.001
P. Robustelli, S. Piana, and D. E. Shaw, Proc. Natl. Acad. Sci. 115 (21) (2018). https://doi.org/10.1073/pnas.1800690115
G. Bussi, D. Donadio, and M. Parrinello, J. Chem. Phys. 126 (1), 014101 (2007). https://doi.org/10.1063/1.2408420
M. Parrinello and A. Rahman, J. Appl. Phys. 52 (12), 7182 (1981). https://doi.org/10.1063/1.328693
J. Tsai, R. Taylor, C. Chothia, and M. Gerstein, J. Mol. Biol. 290 (1), 253 (1999). https://doi.org/10.1006/jmbi.1999.2829
L. Martínez, PLoS One 10 (3), e0119264 (2015). https://doi.org/10.1371/journal.pone.0119264
Python Release Python 3.6.9. https://www.python.org/downloads/release/python-369/
S.-H. Roh, C. F. Hryc, H.-H. Jeong, et al., Proc. Natl. Acad. Sci. 114 (31), 8259 (2017). https://doi.org/10.1073/pnas.1704725114
Funding
This work was supported by the Russian Science Foundation (grant no. 19-74-20055).
Author information
Authors and Affiliations
Corresponding author
Additional information
The original online version of this article was revised due to a retrospective Open Access order.
Rights and permissions
Open Access. This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Panina, I.S., Mamchur, A.A., Yaroshevich, I.A. et al. Study of GroEL Conformational Mobility by Cryo-Electron Microscopy and Molecular Dynamics. Crystallogr. Rep. 66, 846–853 (2021). https://doi.org/10.1134/S1063774521050163
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1063774521050163