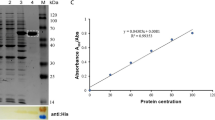
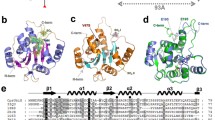
Abstract
Urate oxidase catalyzes the oxidative degradation of uric acid to allantoin via peroxide formation by a radical recombination mechanism. Here the crystal structure of urate oxidase (residues 4-310) from Bacillus subtilis 168 (BsUOX) was solved at 2.6 Å resolution. Both crystal structure and small angle X-ray scattering data confirmed that the BsUOX adopts a tetrameric conformation. Comparative analysis of BsUOX structure alignment with crystal structure of urate oxidase complexed with uric acid from Aspergillus flavus (PDB entry 4D12) showed some conserved BsUOX amino acid residues, Thr69, Ser243, Gln245, and Asn271, in the active site region that can potentially bind uric acid. Residues Ile244 and Gln299 are also predicted to interact with the uric acid via hydrophobic interactions but needs further confirmation. This work will be helpful for further functional and biochemical studies of the enzyme for future drug design and development against gout and hyperuricemia.





Similar content being viewed by others
REFERENCES
M. Oda, Y. Satta, O. Takenaka, and N. Takahata, Mol. Biol. Evol. 19, 640 (2002).
F. Ghaemi-Oskouie and Y. Shi, Curr. Rheumatol. Rep. 13, 160 (2011).
F. Martinon, Curr. Rheumatol. Rep. 12, 135 (2010).
X. Liu, M. Wen, J. Li, et al., Appl. Microbiol. Biotechnol. 92, 529 (2011).
Z. Chen, Z. Wang, X. He, et al., Appl. Microbiol. Biotechnol. 79, 545 (2008).
X. W. Wu, C. C. Lee, D. M. Muzny, and C. T. Caskey, Proc. Natl. Acad. Sci. USA 86, 9412 (1989).
X. M. Wu, D. M. Muzny, C. C. Lee, and C. T. Caskey, J. Mol. Evol. 34, 78 (1992).
J. T. Kratzer, M. A. Lanaspa, M. N. Murphy, et al., Proc. Natl. Acad. Sci. USA 111, 3763 (2014).
F. Dabbagh, M. B. Ghoshoon, S. Hemmati, et al., Curr. Pharm. Biotechnol. 17, 141 (2015).
S. Amaro, D. Soy, V. Obach, et al., Stroke 38, 2173 (2007).
E. W. Kellogg and I. Fridovich, J. Biol. Chem. 252, 6721 (1977).
A. K. Mandal and D. B. Mount, Annu. Rev. Physiol. 77, 323 (2015).
L. X. Chen and H. R. Schumacher, Best Pract. Res. Clin. Rheumatol. 20, 673 (2006).
Y. Y. Sautin and R. J. Johnson, Nucleos. Nucleot. Nucleic Acids 27, 608 (2008).
S. E. Sattui and A. L. Gaffo, Ther. Adv. Musculoskelet. Dis. 8, 145 (2016).
W. M. Gentry, M. P. Dotson, B. S. Williams, et al., Am. J. Health-Syst. Pharm. 71, 722 (2014).
R. Garg, H. R. Sayles, F. Yu, et al., Arthritis Care Res. (Hoboken) 65, 571 (2013).
S. Patel, A. Le, and S. Gascon, Am. J. Health-Syst. Pharm. 69, 1015 (2012).
J. Feng, L. Wang, H. Liu, et al., Appl. Microbiol. Biotechnol. 99, 7973 (2015).
W. Li, S. Xu, B. Zhang, et al., PLoS One 12, e0177877 (2017).
J. Li, Z. Chen, L. Hou, et al., Protein Expr. Purif. 49, 55 (2006).
M. I. Shaaban, E. Abdelmegeed, and Y. M. Ali, J. Microbiol. Biotechnol. 25, 887 (2015).
P. Pfrimer, L. M. de Moraes, A. S. Galdino, et al., J. Biomed. Biotechnol. 2010, 674908 (2010).
A. Vagin and A. Teplyakov, Acta Crystallogr., Sect. D: Biol. Crystallogr. 66, 22 (2010).
A. A. Lebedev, A. A. Vagin, and G. N. Murshudov, Acta Crystallogr., Sect. D: Biol. Crystallogr. 64, 33 (2008).
T. Hibi, Y. Hayashi, H. Fukada, et al., Biochem. 53, 3879 (2014).
G. N. Murshudov, P. Skubak, A. A. Lebedev, et al., Acta Crystallogr., Sect. D: Biol. Crystallogr. 67, 355 (2011).
P. Emsley and K. Cowtan, Acta Crystallogr., Sect. D: Biol. Crystallogr. 60, 2126 (2004).
P. Emsley, B. Lohkamp, W.G. Scott, and K. Cowtan, Acta Crystallogr., Sect. D: Biol. Crystallogr. 66, 486 (2010).
W. L. DeLano, PyMOL (http://www.pymol.org, 2002).
D. Svergun, C. Barberato, and M. H. J Koch, J. App. Crystallogr. 28, 768 (1995).
N. Colloc’h, M. el Hajji, B. Bachet, et al., Nat. Struct. Biol. 4, 947 (1997).
R. D. Imhoff, N. P. Power, M. J. Borrok, and P. A. Tipton, Biochem. 42, 4094 (2003).
S. Bui, D. von Stetten, P. G. Jambrina, et al., Angew. Chem. Int. Ed. Engl. 53, 13710 (2014).
P. Retailleau, N. Colloc’h, D. Vivares, et al., Acta Crystallogr., Sect. D: Biol. Crystallogr. 60, 453 (2004).
L. Holm and P. Rosenstrom, Nucleic Acids Res. 38, W545 (2010).
X. Robert and P. Gouet, Nucleic Acids Res. 42, W320 (2014).
T. Hibi, A. Kume, A. Kawamura, et al., Biochemistry 55, 724 (2016).
ACKNOWLEDGMENTS
We thank the staff of Beam line BL19U1 at SSRF for assistance in data collection. This work was financially supported by the grants from the Ministry of science and technology of China (2016YFA0500700), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDPB10, XDB08010101), and the National Natural Science Foundation of China (grant 31330018, 31770805, and 91540103).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Nayab, A., Moududee, S.A., Shi, Y. et al. Crystal Structure of Urate Oxidase from Bacillus Subtilis 168. Crystallogr. Rep. 64, 1126–1133 (2019). https://doi.org/10.1134/S1063774519070149
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1063774519070149