Abstract
Plants have a unique ability to adapt ontogenesis to changing environmental conditions and the influence of stress factors. This ability is based on the existence of two specific features of epigenetic regulation in plants, which seem to be mutually exclusive at first glance. On the one hand, plants are capable of partial epigenetic reprogramming of the genome, which can lead to adaptation of physiology and metabolism to changed environmental conditions as well as to changes in ontogenesis programs. On the other hand, plants can show amazing stability of epigenetic modifications and the ability to transmit them to vegetative and sexual generations. The combination of these inextricably linked epigenetic features not only ensures survival in the conditions of a sessile lifestyle but also underlies a surprisingly wide morphological diversity of plants, which can lead to the appearance of morphs within one population and the existence of interpopulation morphological differences. The review discusses the molecular genetic mechanisms that cause a paradoxical combination of the stability and lability properties of epigenetic modifications and underlie the polyvariance of ontogenesis. We also consider the existing approaches for studying the role of epigenetic regulation in the manifestation of polyvariance of ontogenesis and discuss their limitations and prospects.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
In 2022 will be the 150th anniversary of the birth of Nikolai Konstantinovich Koltsov, the forerunner of many fundamental discoveries and scientific directions of the 20th century, including epigenetics (Morange, 2011; Ramenskii, 2018). Among the predictions of the great scientist, we can find not only arguments for epigenetic phenomena but also the idea of the influence of methylation on the manifestation of hereditary traits. In 1915, he argued that “in any organic compound, a hydrogen atom can be abruptly replaced by a group of CH3” (Koltsov, 1915). Later, the idea about the influence of nongenetic factors on the implementation of the genotype, about the multiplicity of “epigenetic trajectories” possible for one genotype, was developed by Conrad H. Waddington (1968), and the discovery of the role of DNA methylation and histone proteins in the epigenetic regulation of gene action fully confirmed the prediction of N.K. Koltsov. Today, thanks to significant advances in the study of molecular genetic mechanisms of epigenetic changes, epigenetics has become one of the most important areas of genetics, developmental biology, and molecular biology. Although the concept of epigenetics is interpreted differently in different sources (see the review of Tikhodeev, 2015), most researchers agree that epigenetic changes are modification changes that are not related to changes in the DNA sequence but are capable of being preserved in several cell generations even in the absence of factors that caused these changes (Jablonka and Raz, 2009; Ptashne, 2013; Noble, 2015). Inherited changes in the expression of gene alleles caused by epigenetic modifications (EM) are called epialleles.
Epigenetic mechanisms of regulation of gene expression underlie several biological processes that ensure the existence and development of plants and animals (Allis et al., 2010). The functioning of the chromosomal apparatus during mitosis and meiosis is impossible without maintaining the heterochromatin state of the centromeric and telomeric regions. The condition of chromatin, which is determined by the presence of “epigenetic marks” (modified versions of histone proteins and methylcytosine in DNA), affects the processes of repair and transcription (Razin and Bystritskii, 2009). EM protect the integrity of the genome by suppressing the transcription of genes of mobile elements and the possibility of their movement and reproduction (Fultz et al., 2015). Epigenetic inactivation (silencing) of duplicated genes after allo- and autopolyploidy and genomic duplications provides homeostasis and opens up opportunities for the evolution of gene function (Song and Chen, 2015). The dose compensation of sex chromosome genes, which has been well studied in animals, also occurs in dioecious plants with sex chromosomes and is also based on EM (Muyle et al., 2017, 2018).
EM plays a key role in the development processes. The transition to a new stage of ontogenesis and the development of each new structure are associated with local epigenetic reprogramming, i.e., the suppression of the expression of genes that are active at the previous stage, and, at the same time, the activation of a new group of genes that determine the specifics of the new organ/stage (Xiao and Wagner, 2015; Xu et al., 2018; Yan et al., 2020). The established profile of gene expression is maintained by descendant cells. It is cellular memory, based on epigenetic mechanisms, that underlies the maintenance of structural and functional features of all tissues and organs of multicellular organisms during ontogenesis (Nashun et al., 2015; Birnbaum and Roudier, 2016).
Epigenetic processes that are an integral part of the genetic program of development and are initiated by internal signals are called constitutive epigenetic control of development (Bräutigam and Cronk, 2018). When the function of key epigenetic regulators is impaired, plant cells lose the ability to maintain cellular identity (differentiated state) and acquire callus characteristics and the capability of somatic embryogenesis (Aichinger et al., 2009; Chen et al., 2010; Bratzel et al., 2010).
All the mentioned functions of EM are common to animals and plants. At the same time, EM in plants perform an additional function of adaptation to local environmental conditions and the effects of adverse factors due to changes in development programs. The interaction between development and adaptive (stress) response programs allows plants to quickly find the optimal life strategy in a limited resources (Herms and Mattson, 1992; Huot et al., 2014). This is possible due to the fact that various unfavorable conditions (lack of light, water, nutrition, hypo- or hyperthermia, exposure to pathogens, etc.) can initiate partial epigenetic reprogramming of the genome, which opens up the possibility for changing the development program. Epigenetic correction of a genetically determined development program in response to the action of external signals is called facultative epigenetic development control (Bräutigam and Cronk, 2018).
The possibility of changing the development program is fundamentally important in the conditions of an attached or passive (in aquatic plants) lifestyle and causes amazing plasticity of ontogenesis, which is characteristic of plants (Serebriakov and Serebriakova, 1972; Zhukova and Glotov, 2001; Kalinkina, 2017; Sultan, 2017; Notov and Zhukova, 2019). Moreover, in plants, phenotypic changes caused by external influences can be inherited by vegetative and sexual progeny (Hauser et al., 2011; Heard and Martienssen, 2014; Lämke and Bäurle, 2017; He and Li, 2018). This review will focus on the role of EM in the manifestation of this amazing ability to reprogram ontogenesis and analyze the mechanisms that can initiate epigenetic reprogramming of ontogenesis and the appearance of morphotypes under the influence of environmental signals.
FEATURES OF PLANT BIOLOGY UNDERLYING THE PLASTICITY OF DEVELOPMENT
Plasticity of development is the most important condition for survival in the situation of an attached or a passive (in the case of aquatic plants) lifestyle. It is caused by the ability of plant cells to change their identity in response to signals from surrounding cells, physiological signals, and signals from the environment. This ability, that was discovered in the studies with chimeric plants (Poethig, 1989; Irish, 1991; Szymkowiak and Sussex, 1996) and in experiments on de novo regeneration of roots (reviews Xu, 2018; Li, 2021) and shoots (Shin et al., 2020; Lardon and Geelen, 2020) from various organs indicates that plant cells easily change the genetic program of development.
In a plant, the formation of new organs occurs throughout ontogenesis, so changes in the program of ontogenesis can occur at any stage of the life cycle, including in an adult plant. These changes are facilitated by the fact that they can affect not the entire body but its parts because plants have a modular structure (Hallé, 1986; Oborny, 2019). Modules are formed sequentially as a result of the functioning of the apical meristems of the shoot and root. The module of a shoot is an internode with a leaf and an axillary meristem. The root module is a fragment with a root and a lateral root extending from it. When conditions (external or internal) change the fate of some, the youngest modules may change. Such a local rearrangement of morphogenesis can ensure the adaptation of the formed parts of the plant to new conditions without changing the entire structure of the plant.
Thanks to epigenetic mechanisms, the new program can be maintained not only by a cell clone but also by vegetative generations since many plants are capable of vegetative reproduction. A lot of experimental data about the inheritance by vegetative offspring of the DNA methylation pattern, which is caused by stress effects on maternal plants, has been obtained. In the white clover Trifolium repens (Leguminósae), changes in DNA methylation caused by drought stress on the mother plant were preserved in five studied vegetative generations (Rendina González et al., 2018). The specific methylation pattern of the invasive clonal plants of alligator weed, Alternanthera philoxeroides (Amaranthaceae) from different ecological and geographical places in China was maintained when transferred to common conditions for 2–3 generations, although some of the EM (38%) were preserved for ten vegetative generations (Shi et al., 2019).
Stable transmission of individual epialleles to offspring is also shown for many species with obligate generative reproduction: in toadflax Linaria vulgaris, arabidopsis Arabidopsis thaliana, maize Zea mays, rice Oryza sativa, tomatoes Solanum lycopersicum, etc. (Cubas et al., 1999; Kakutani, 2002; Manning et al., 2006; Miura et al., 2009; O’Malley and Ecker, 2012; Zhang et al., 2012; Weigel and Colot, 2012; Johannes and Schmitz, 2019). Some epialleles in plants are so stable that they demonstrate monogenic inheritance and it is difficult to simply distinguish them from monogenic mutations (Bondada et al., 2020).
Global genome demethylation is typical for animal gametogenesis and early embryogenesis, its absence contributes to the transmission of EM to sexual offspring in plants (Pikaard and Scheid, 2014). This feature and the molecular mechanisms of EM maintenance operating in plant cells (see below) explain the surprisingly high proportion of stably inherited methylcytosine sites (91%) and methylated regions (99.998%) in genetically homogeneous Arabidopsis inbred lines (MA lines), which were obtained by self-pollination over 30 generations from a single founder plant (Schmitz et al., 2011; Hofmeister et al., 2017).
The absence of early separation of cells of the germ line is an important feature of plants. In plants, generative cells are formed from the initial cells of the germ line, which differ from the surrounding somatic meristem cells only by their position in the apex of the shoot but not by the nature of determination, and, in case of death, they can be replaced by somatic cells (Whipple, 2012; Pikaard and Scheid, 2014). Therefore, the genetic and epigenetic information of somatic cells can potentially be passed on to offspring.
Thus, the characteristics of plants determine the possibility of their survival with an sessile/passive lifestyle. By changing the genetic program of development, plants can adapt ontogenesis to environmental conditions, showing plasticity of development. A new variant of ontogenesis can be maintained due to epigenetic mechanisms and even transmitted to vegetative and sexual offspring. Due to these specific features, the variability of the plant phenotype can take the form of polyvariance, i.e., the existence of certain morphotypes based on a single genotype.
HOW EPIGENETIC MODIFICATIONS CAN AFFECT THE PHENOTYPE
The main mechanism of epigenetic modifications in plants is DNA methylation and modification of histone proteins. Epigenetic phenomena also include the so-called protein inheritance, i.e., the inheritance by a cell clone of an altered conformation of protein molecules, described in yeast, animals, and humans (Harvey et al., 2018). Prion-like proteins are also found in plants. However, they do not normally form aggregates in cells; this is prevented by plant metabolites (Surguchov et al., 2019).
Three groups of genes provide EM of DNA and histones (Pikaard and Scheid, 2014; Allis and Jenuwein, 2016). The genes of the first group (writers) encode enzymes that carry out modifications of DNA (cytosine methylation) and histones (methylation, acetylation, ubiquitination, phosphorylation, histone sumoylation, etc.). The genes of the second group (erasers) perform the opposite function, removing these marks. The genes of the third group (adaptor proteins or readers) recognize epigenetic marks and affect a variety of cellular processes through interaction with other proteins (Fig. Suppl see https://doi.org/ 10.1134/S1062360421060047). Some proteins can combine different functions. For example, CMT chromometylases are proteins of both the first and third groups since they recognize histone repressive marks and methylate DNA in the in close proximity (Zhang et al., 2018). Similarly, histone H3K9 methyltransferases perform their main function after the recognition of methylcytosine 5mC (Li et al., 2018). However, this classification helps to systematize extensive information on epigenetic regulation.
Group one: writers. This group includes genes encoding DNA-methyltransferases and histone-methyltransferases. Unlike animals, in which methylated cytosine is usually guanine-adjacent cytosine (CG sites), in plants, cytosine in any surroundings can be methylated (CG, CHG, and CHH, where H is A, C, or T). This is possible due to the existence of a unique family of CHROMOMETHYLASE (CMT) genes, as well as genes that control the RNA-dependent DNA methylation pathway, or RdDM (RNA-directed DNA Methylation pathway). The RdDM pathway is based on the mechanism of RNA interference, which involves variants of the conservative Dicer and Risc complexes (including the DCL and AGO proteins, respectively) as well as plant-specific RNA polymerase complexes IV and V (Zhang et al., 2018; Raju et al., 2019; Gallego-Bartolomé, 2020).
The effect of DNA methylation on gene expression depends on the context in which cytosine is methylated, and on the location of methylation sites. Cytosine methylation in the regulatory parts of the gene is most often associated with a decrease in the level of transcription (Vaniushin, 2006; Law and Jacobsen, 2010). However, in some cases, due to interaction with proteins from group three, the effect may be the opposite (Harris et al., 2018). Histone methylation is associated with either activation or repression of genes depending on the position and number of methyl groups. H3K4me3, H3K9me3, and H3K36me3 correlate with transcription activation, while the presence of the H3K27me3 marks genes with a reduced transcription level (Zhang et al., 2007, 2009; Roudier et al., 2011). The H3K9me2 and H3K27me1 marks are usually localized in the centromeric regions of chromosomes and are typical for repeats and mobile elements that also contain highly methylated DNA (Bernatavichute et al., 2008; Zhang et al., 2009; Roudier et al., 2011).
Histone methylation is carried out by several families of histone methyltransferases, which methylate different lysine residues and work as part of different protein complexes, among which PRC1 and PRC2 complexes are the most studied (Liu et al., 2010). These complexes include both histone deacetylases (present in PRC1, PRC2) and histone ubiquitinases (PRC1), which also cause chromatin compactification and gene repression (Derkacheva and Hennig, 2014; Mozgova and Hennig, 2015; Xiao and Wagner, 2015; Förderer et al., 2016).
Histone acetylases loosen chromatin and activate gene expression; ubiquitination has either an activating or repressing effect on the state of chromatin. Other modifications of histone proteins can also affect the state of chromatin (Berger, 2007; Fenley et al., 2018), although their participation in epigenetic processes is less studied. Histone acetylases and ubiquitinases can also work as part of various protein complexes, including complexes that maintain the active state of chromatin (Fletcher, 2017; Ornelas-Ayala et al., 2021).
Group two: erasers. Active removal of methyl groups from cytosine in plants is provided by plant-specific enzymes with glycosylase activity; they remove methylated cytosine and initiate its replacement with unmethylated cytosine (Li et al., 2018; Parrilla-Doblas et al., 2019; Liu and Lang, 2019). The removal of acetyl and methyl groups from histone proteins is provided by several families of histone deacetylases (Ma et al., 2013; Chen et al., 2020) and demethylases (Prakash et al., 2014), among which there are plant-specific ones.
Group three: readers recognize (“read”) the installed marks or the absence of marks. This is the most diverse group of epigenetic regulators (ER), which also most often act as part of protein complexes (Torres and Fujimori, 2015; Grimanelli and Ingouff, 2020). Readers often read different histone labels at the same time, including phosphoric and ubiquitin marks. The epigenetic regulators of this group often include chromatin proteins that can recognize EM and affect the noncovalent interaction of DNA with histone proteins, change the packing density and localization of nucleosomes, and also catalyze the inclusion of histone variants (Li et al., 2007; Narlikar et al., 2013).
Group three proteins have not only binding sites with DNA methylcytosine or histone tags but also binding sites with nonhistone proteins. Because of this, readers provide interaction between epigenetic marks (modifications of histones and DNA) and other cellular processes in which a nonhistone protein is involved (Li, Y. and Li, H., 2012; Patel, 2016; Grimanell and Ingouff, 2020). For example, readers can interact with proteins that establish or remove marks from DNA and histones, contributing to the spreading or elimination of marks (Grimanell and Ingouff, 2020; Scheid et al., 2021). They can contribute to the opening of chromatin, due to the inclusion of histone variants that change the state of chromatin (Kouzarides, 2007; Sijacic et al., 2019). They can directly affect transcription by interacting with transcription factors (TF) or their cofactors or they have a DNA-binding domain and act as TF pioneers, the affinity and specificity of which depend on the presence of methylcytosine in the DNA (Zhu et al., 2016; Kribelbauer et al., 2019; Grimanell and Ingouff, 2020).
Group three proteins can influence the choice of promoters (Le et al., 2020) as well as posttranscription events (Fig. Suppl see https://doi.org/10.1134/ S1062360421060047). By interacting with proteins of the RNA polymerase complex, proteins that control the maturation of pre-mRNA (proteins involved in polyadenylation, splicing) readers determine the choice of splicing sites (Ullah et al., 2018; Wei et al., 2018; Wu et al., 2020) and polyadenylation sites (Duan et al., 2017; Zhang et al., 2021). This means that readers are participants in the regulation of gene expression at different stages of the implementation of genetic information, combining the epigenetic marks of DNA and histones with the basic mechanisms of regulation of gene expression. In fact, the epigenome determines which genes are transcribed in a given tissue, which of the alternative promoters, splicing sites, and polyadenylation sites will be selected in a given cell clone, and therefore which of the protein isoforms will function in a given tissue at a given stage of ontogenesis and under given environmental conditions. Thus, EM not only preserve the expression profile that is established as a result of the action of TF but also participate in its establishment at all stages of mRNA formation and maturation.
The considered three groups of genes control EM not only in plants but also in animals. However, the regulation of the expression of these groups of genes in plants should have its characteristics. On the one hand, the activity of these genes should depend on environmental conditions. In other words, plants should have mechanisms that can initiate partial epigenetic reprogramming of the genome and the establishment of a new development program in changed environmental conditions. On the other hand, plants should have the ability to preserve a new adaptive development program and pass it on to their descendants. This paradoxical combination of stability and lability of EM indicates the existence of special mechanisms in plants that control the dynamics of EM. Let’s consider the available data that can explain, at least partially, the unique features of epigenetic regulation in plants.
MOLECULAR MECHANISMS UNDERLYING THE INHERITANCE OF EPIGENETIC MODIFICATIONS IN PLANTS
EM are different in the stability of inheritance through sexual generations and, as a rule, do not obey the Mendelian laws. Nevertheless, heritability, at least in the form of “heritable changeability” (Le Goff et al., 2021) is a characteristic feature of EM, which is specific for plants.
The ability of EM to persist during meiosis is partly explained by the peculiarities of plant biology (see above). However, there are also molecular mechanisms that can support this ability. Their effect depends on the type of epiallele (“silent” hypermethylated allele or actively working hypomethylated epiallele), on the copy number variation of genes, on the presence of special motifs in the loci that attract DNA methyltransferases or other ER, on the number of DNA methylation sites, their type (CG, CHG, CHH) and localization in the gene, and on the presence of histone modifications and their type (Catoni et al., 2017; Li et al., 2020; Williams and Gehring, 2020).
These dependencies are complex and are not fully investigated. Nevertheless, the main tools for the preservation of EM are maintenance DNA-methyltransferases, which are also present in animals, as well as plant-specific mechanisms of positive feedback loop between chromatin marks and methylcytosine marks (5mC). When maintaining the established epigenetic profile, CMT chromometylases methylate DNA, focusing on the presence of H3K9me2 chromatin marks, while histone methyltransferases KYP/SUVH4, etc. are focusing on the presence of 5mC (Baubec et al., 2010; Du et al., 2015; Stoddard et al., 2019). This mechanism explains the stable “silence” of hypermethylated alleles (for example, in the Arabidopsis SUP gene).
There is positive feedback loop during establishing methylation involving small RNAs de novo (RdDM pathway). Group three proteins (readers) play an important role in this multicomponent process. Some readers recognize chromatin labels of methylated H3K9me2 and direct the work of RNA polymerase IV, which is involved in the formation of small RNAs. Others recognize 5mC and attract RNA polymerase V, which, through intermediaries (primarily the AGO protein with a small RNA loaded into it), can attract a specific de novo DRM2 DNA methyltransferase to the DNA sites from which pre-mRNA is read (Matzke and Mosher, 2014; Erdmann and Picard, 2020). Thanks to these mechanisms, the EM that appeared during reprogramming of the genome will be maintained until the moment when other regulators interfere in the process, which can be activated by changing environmental conditions (see below). Activating H3K4me3 and H3K18ac histone marks interfere with the RdDM pathway and attract DNA demethylase/glycosylase ROS1, which removes 5mC and creates conditions for stable maintenance of hypomethylated epialleles (Li et al., 2020).
The resulting small RNAs, which are constantly generated during a positive feedback loop, can move in the plant via plasmodesma and phloem (Tamiru et al., 2018; Pagliarani and Gambino, 2019), and possibly be transmitted to descendants, as is shown for the nematode C. elegans (Xu et al., 2018). No clear evidence has yet been obtained for the participation of small RNAs caused by stress on parents in the transmission of epigenetic information to offspring in plants (Morgado et al., 2017; Colicchio et al., 2020). Nevertheless, the resulting mobile small RNAs can penetrate germinal cells (Duempelmann et al., 2020) and initiate DNA methylation and subsequent histone modifications in them, which can be transmitted to descendants and lead to the inheritance of phenotypic changes: ontogenetic or physiological and biochemical changes associated with resistance to the stress experienced by parent plants (Tamiru et al., 2018; Liegard et al., 2019).
The maintenance of epigenetic marks in certain gene loci in a series of generations can be facilitated by repeats, which often represent to some extent degraded mobile elements embedded in the regulatory region of genes. Thus, the presence of repeats in the regulatory region of the Arabidopsis FWA gene and maize B1 gene explains the frequent occurrence of epialleles of these genes (Henderson and Jacobsen, 2007). The stability of the EM of such genes is indicated by the fact that several varieties of economically valuable crops have been created based on similar EM (review Lisch, 2013).
In mammalian genomes, nucleotide motifs that promote the attraction of DNA methyltransferases to the regulatory sites of certain loci were found (Handa et al., 2005; Wienholz et al., 2010; Wang et al., 2019). Plants also have them, although they are less studied (Gouil and Baulcombe, 2016). The peculiarities of the structure of loci, which make them favorite targets of ER regulators, can explain the repeatedly observed cases of stable epialleles of some genes. However, knowledge about the features of DNA or chromatin that can turn loci into “hot” and “cold” EM points is not sufficient. For example, it is still not clear why such a “hot spot” is the SUP arabidopsis (Arabidopsis thaliana) gene, that sets a boundary between male and female reproductive organs in the flower (Bowman et al., 1992). Hypermethylated “silent” epialleles in this gene have been isolated after exposure to neutrons, X-rays, ethylmethyl sulfate, and other chemical mutagens and insertion T-DNA mutagenesis (Jacobsen and Meyerowitz, 1997; Rohde et al., 2002), against the background of genome demethylation in transgenic lines with the suppressed activity of the DNA methyltransferases gene MET1 (Kishimoto et al., 2001) as well as spontaneous demethylation (Bondada et al., 2020). Moreover, epialleles of the SUP gene with different degrees of expressivity and penetrance were found in 11 (of 1028) Arabidopsis ecotypes from a variety of habitats (Bondada et al., 2020).
The maintenance of EM in certain loci may be caused by the presence of cis-elements for binding TF in their regulatory regions, which attract ER (for example, DNA methyltransferases or demethylases), maintaining a hypermethylated or, vice versa, a hypomethylated state of the loci. Long noncoding RNAs (lncRNAs) can also attract ER to certain loci. This is shown for COLDAIR lncRNA, which epigenetically suppresses the Arabidopsis FLC gene that controls the need for vernalization (Kim et al., 2017; Wu et al., 2020), as well as for AG-incRNA4 lncRNA, which causes silencing in the vegetative part of the shoot of the AG gene that controls the development of reproductive organs in the flower (Wu et al., 2018). Due to interaction with ER, TF and lncRNA can cause EM in target genes that have either cis-elements for attracting TF or sites of homology with lncRNA. Such EM effectors, the expression of which is activated under certain conditions (or under stressful influences) can, apparently, play an important role both in the initiation of EM under the influence of new conditions/stresses and in keeping the emerging epialleles in a row of generations while maintaining changed conditions (for example, in 11 generations of inbred rice lines grown in drought conditions) (Zheng et al., 2017).
Clear evidence of the inheritance of EM in plants is the inheritance of 99.998% of methylation regions over 30 generative generations under unchanged environmental conditions established during the study of Arabidopsis inbred lines (MA lines obtained from self-pollination of one plant of the Col ecotype) (Hofmeister et al., 2017). This stability made it possible to create an epigenetic map based on the analysis of the F2 generation from the hybridization of two MA lines and to show that over half of the newly formed epialleles segregated in a Mendelian manner (Hofmeister et al., 2017).
MOLECULAR MECHANISMS THAT INITIATE CHANGES IN THE DEVELOPMENT PROGRAM
Many stress factors (both abiotic and biotic) are among the factors that affect the functioning of the ER and initiate partial reprogramming of the genome. They cause changes in metabolic processes and the appearance of reactive oxygen species (ROS) and nitrogen species, which change the redox balance of the cell and activate redox-dependent signaling systems in cells (Foyer and Noctor, 2013; Locato et al., 2018). The consequence of these processes is a change in the content of donor molecules of methyl groups necessary for the modification of DNA and histones (primarily, S-adenosyl methionine), as well as the effect of ROS and metabolites on the regulatory proteins of epigenetic modifications (Locato et al., 2018; Saravana Kumar et al., 2020; Lindermayr et al., 2020). DNA-demethylating enzymes (Couturier et al., 2013; Duan et al., 2015) and proteins involved in RNA-dependent DNA methylation (Charbonnel et al., 2017; Seta et al., 2017) are among them. The effect of the redox potential and nitric oxide, which is also a powerful oxidant, on proteins involved in DNA methylation and histone protein modification is described (reviews by Locato et al., 2018; Saravana Kumar et al., 2020; Lindermayr et al., 2020). Since cytosine can be methylated in any sequence context in plants, they are particularly sensitive to metabolic changes that cause a decrease in the level of methyl group donors in cells (Lindermayr et al., 2020).
Recently, an additional mechanism of the relationship between stress effects and genome reprogramming has been identified. It has been shown that SUMOylation, a reversible posttranslational modification of various proteins, plays an important role in the activation of the stress response (heat shock, hypothermia, drought, phosphate deficiency, pathogen attack) (Jmii and Cappadocia, 2021). The most important target of the Sumo E3 ligase SIZ1 is the DNA glycosylase ROS1, which is stabilized as a result of this modification and performs DNA demethylation. Mutations in the SIZ1 gene in Arabidopsis lead to an increase in the level of DNA methylation due to a decrease in the content of the ROS1 protein (Kong et al., 2020). Protein SUMOylation is a dynamic and reversible process that is activated by stress influences. Therefore, under stress conditions, the SUMOylation of proteins through the influence on DNA demethylation can play an important role in the reprogramming of the plant genome.
Environmental conditions/stresses can change the activity of ER at both the posttranslational and transcriptional levels. Many ER are transcribed peculiarly, only under certain conditions/stressful influences, or in certain tissues and organs at certain stages of ontogenesis. Transcription specificity has been shown for DNA-methyltransferases and other ER of different plant species (Rohini et al., 2014; Guo et al., 2020; Yu et al., 2021). Moreover, among ER that belong to the same class (for example, DNA demethylases/glycosylases), there are also some differences between target genes, which may be explained by the presence of specific motives in the targets that attract ER or by the fact that TF or lncRNAs that interact with a certain set of target genes are involved in attracting target genes (see above). We should note that there are also components of hormonal signaling pathways among the TF that interact with ER. By attracting ER that change the state of chromatin of their target genes, TF can either activate (Wu et al., 2015) or suppress gene expression (Hasegawa et al., 2018) in response to the action of hormones.
Most of the mentioned factors that contribute to changing the development program when environmental conditions change, act to some extent on epigenetic processes in animal cells. At the same time, there is new information about the specificities of the action of some ER in plants as well as about the discovery of new plant-specific ER. 4 Arabidopsis genes that arose as a result of the domestication of transposons of the Harbinger family are among them. The products of two such genes HDP1 and HDP2 work as part of the IDM1 protein complex, which has histone-acetyltransferase activity and, at the same time, participates in the attraction of DNA demethylase/glycosylase ROS1 (Duan et al., 2016). The products of two other genes, ALP1 and ALP2, also act together and prevent the PRC2 complex from performing repressive H3K27me3 methylation of histones (Liang et al., 2015; Velanis et al., 2020).
The ICU11 and CP2 genes found in Arabidopsis also interact with PRC2-complex proteins (Mateo-Bonmatí et al., 2018; Bloomer et al., 2020). However, this interaction leads to an increase in the effectiveness of epigenetic suppression of target genes. It is assumed that these genes encoding 2-oxoglutarate/Fe(II)-dependent dioxygenases help to remove activating H3K4me2/3 and H3K36me3 marks from histones, which facilitates the further establishment of repressive marks (Bloomer et al., 2020). Recently, the CFK1 gene, encoding a protein with an F-box, which interacts with de novo DNA methyltransferase and starts the process of its degradation, was found in Arabidopsis (Chen et al., 2021).
In addition to these new genes that interfere with the work of already described ER, genes that are representatives of well-known families of epigenetic regulators but perform a function opposite to that performed by their paralogs are described in plants. Many ER in plants are presented by several genes, which is due to the duplication of the genome characteristic for plants. The function of not all representatives has been analyzed, and their study brings surprises. For example, the SUVH family of histone H3K9 methyltransferases (SU(VAR)3-9 HOMOLOG) in Arabidopsis is represented by nine paralogs, among which SUVH4/KYP, SUVH5, and SUVH6 provide repressive methylation of H3K9. Due to the presence of the SRA domain, they recognize methylated DNA and methylate histones near these sites (Li et al., 2018). At the same time, SUVH1, SUVH3, SUVH7, and SUVH8 perform the opposite function. They have lost the methyltransferase domains, but they recognize methylated DNA in the promoter region of DNA demethylase/glycosylase ROS1 due to the SRA domain. Interacting with transcription activator proteins with the DnaJ domain, the SUVH1, SUVH3, SUVH7, and SUVH8 proteins activate the transcription of ROS1 and subsequent demethylation of DNA (Xiao et al., 2019). There are likely to be more examples of such subneofunctionalization with the increase of our knowledge about the function of ER paralogs in plants.
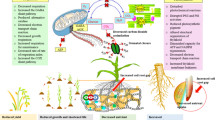
In conclusion of this part, we note that environment-induced partial reprogramming of the genome can lead to activation genes for a protective response and/or change in the plant’s (its growing modules) ontogeny program. A change in the development program of a particular module or organ is realized at the early stages of its ontogenesis, most often in the cells of the apical meristems of the shoot and root, which contain stem (pluripotent) cells and give rise to all organ primordia. However, the activation (or reactivation) of the development program in plants can be observed even in mature organs (Ezhova and Kupriianova, 2019). This is explained by the fact that pluripotent cells in plants can appear de novo due to the removal of epigenetic repression from the stemness genes, which is maintained by numerous ER in differentiated tissues (Albert and Ezhova 2013; Gaillochet and Lohmann, 2015). The removal of epigenetic repression from genes of stemness can cause the development of additional blades and comb-like outgrowths even on mature plant leaves (Fedotov et al., 2017; Kupriyanova et al., 2021). Ontogenetic reversals caused by weather conditions also reflect the possibility of restarting genetic development programs due to the renewal of the stem cell pool (Fig. 1).
Ontogenetic reversions in calendula Calendula officinalis (this variety was created by V.V. Sakharov at the Koltsov Institute of Developmental Biology of the Russian Academy of Sciences) and water avens Geum rivale in the conditions of a wet, cold summer of 2019. On one the same plant of calendula, along with inflorescences with normal ontogenesis (a), inflorescences forming new apical meristems, which also form inflorescences and seeds (b, c) can be observed (c—capitulum of the first order with ripe seeds (at the bottom); 15 additional inflorescences of the second order formed by it, which are at different stages of ontogenesis (at the top)). (d) On the same plant of the water avens with normal flower ontogenesis, (e, f) there are flowers in the center of which new shoots with inflorescences are formed. Photo by the author.
MAIN EXPERIMENTAL APPROACHES FOR STUDYING THE ROLE OF EPIGENETIC MODIFICATIONS IN THE POLYVARIANCE OF DEVELOPMENT
The plasticity of plant development is manifested in a pronounced variability of traits in plants of the same species. Stochastic fluctuations in the level of expression, as well as changes in gene expression caused by external and internal factors that change development, are not always associated with EM. To prove the causal relationship between EM and the phenotype, it is necessary to be sure that there is no connection between the phenotype and genetic polymorphism and to distinguish temporary changes in gene expression that do not persist during cell division from long-lasting EM. There are several approaches for studying the role of EM in the manifestation of polyvariance of development that have different degrees of evidence (Table 1, see explanations in the Suppl, https://doi.org/10.1134/S1062360421060047).
However, as the knowledge about the genetic control of ontogenesis deepens, new opportunities will appear related to testing the expression level and methylation profile not of the entire genome but of key conservative genes, changes in the work of which can potentially cause a similar phenotype. An example of such studies is the analysis of the effect of water deficiency on the development of stomata on the leaves of Arabidopsis plants (Tricker et al., 2012). It is shown that, under these conditions, hypermethylation of the SPCH gene, which controls the development of stomata, was observed. This led to a decrease in their number on the leaf. A high level of DNA methylation of the SPCH gene and a reduced number of stomata were inherited by descendants (Tricker et al., 2013).
For now, the leaf development program is one of the most fully studied and conservative genetic programs (Fritz et al., 2018; Satterlee and Scanlon, 2019)). In this regard, very good prospects are opened up for studying the genetic and epigenetic foundations of heterophylly. The analysis of the expression of genes that play a central role in conservative genetic programs of leaf development was carried out on two species of heterophilic plants (Nakayama et al., 2014; Li et al., 2017; Kim et al., 2018). In plants Rorippa aquatica (Brassicaceae) (Nakayama et al., 2014) and Hygrophila difformis (Acanthaceae) (Li et al., 2017), the dissection of submerged leaves appeared to be a result of activating highly conservative genes in the leaf primordia that support the pluripotency of leaf cells. These are genes of the KNOX I class, the expression of which supports cell division and determines the possibility of complication of the leaf structure (Blein et al., 2008). In both species, these genes are not active in the terrestrial whole-edged leaves but are expressed during the development of dissected leaves (Nakayama et al., 2014; Li et al., 2017). Although these studies do not analyze DNA methylation or histone modification in leaves of different types, it can be assumed that it is the epigenetic regulation of KNOX I genes under the influence of environmental conditions that underlies heterophylly. KNOX I genes are a well-known target of epigenetic regulation (Lodha et al., 2008; Gaillochet and Lohmann, 2015). Therefore, there is no doubt that in the regulatory regions of the KNOX I genes of these species, repressive marks in terrestrial leaves and their absence in submerged ones can be found. In these studies, it was shown that the main external factor causing a change in the morphology of leaves is temperature. Lowering the temperature from 26 to 20°C is enough for the plant to begin to form dissected leaves instead of whole ones. Therefore, R. aquatica and H. difformis are ideal models for future studies of the dynamics of epigenetic modifications in KNOX I-genes under changing external conditions. These models may allow us to decipher the sequence of molecular events that are triggered by external influences and directly or indirectly activate or block the activity of genes involved in the epigenetic regulation of KNOX I genes.
CONCLUSIONS
Due to the use of various approaches to studying the role of epigenetic mechanisms in the polyvariance of plant development, the following statements have been experimentally proven today: (1) environmental conditions, stressful effects cause epigenome changes; some of the emerging EM are adaptive; (2) epigenetic changes are inherited while maintaining the conditions that caused the changes; over half of the EM is stably transmitted to descendants in accordance with Mendelian laws; (3) when the conditions change, the previous EM gradually disappear, although the level of stability of different EM is different; (4) intraspecies morphological variability of plants is caused not only by genetic polymorphism but also by epigenetic differences between plants; (5) many morphological, physiological, and biochemical features that distinguish different parts of the same plant are the result of facultative epigenetic mosaicism, which occurs due to differences in microconditions affecting different parts of the plant.
These statements indicate the existence of unique features of the functioning of epigenetic mechanisms in plants. Indeed, the functioning of ER in plants is under the control of environmental conditions. The environmental impact may be nonspecific (due to a decrease in the level of methyl group donors) or specific, which is based on the activation of the expression of individual ER genes and their effect on certain target genes. These and other mechanisms considered in the review may cause changes in the expression of certain groups of genes and explain the adaptivity of EM. The preservation of adaptive EM in offspring is ensured by maintenance methyltransferases and positive feedback loops between different epigenetic marks. Recently discovered plant-specific genes can also influence the work of already known ER. The products of these genes can either disrupt or activate the function of ER. There is no doubt that new ER that affect the dynamics of EM will be discovered in plants since the function of ~30% of the genes of the nuclear genome is still unknown even in Arabidopsis.
There are more and more studies of the epigenetic bases of intraspecies phenotypic variability; however, even with the “omics” execution, additional confirmation is required that the identified associations between the phenotype and the epialleles of candidate genes are due to “cause-and-effect” relationships. Today, new approaches are being developed for such confirmations. Using special systems for epigenetic editing of genomes, it is possible to use them to attract ER proteins to candidate genes and to study the effect of these modifications on the phenotype (Gallego-Bartolomé et al., 2020; Fal et al., 2021; Selma and Orzáez, 2021). Omics approaches and biotechnologies of directed control of epigenetic processes open up new prospects in the study of the scientific foundations of the plasticity of plant development and the practical use of inherited EM.
REFERENCES
Aichinger, E., Villar, C.B.R., Farrona, S., et al., CHD3 proteins and Polycomb group proteins antagonistically determine cell identity in Arabidopsis, PLoS Genet., 2009, vol. 5, art. e1000605.
Albert, E.B. and Ezhova, T.A., Genetic regulation of plant shoot stem cells, Russ. J. Genet., 2013, vol. 49, no. 2, pp. 127–140.
Allis, C.D. and Jenuwein, T., The molecular hallmarks of epigenetic control, Nat. Rev. Genet., 2016, vol. 17, no. 8, pp. 487–500.
Allis C.D., Jenuwein T., Reinberg D. (Eds.) Epigenetics. New York: Cold Spring Harbor Laboratory Press, 2007. (Russ. ed.: Ellis S.D., Dzhenyuveyn T., Reynberg D. (red.) Epigenetika. Moscow: Tekhnosfera Publ., 2010. (in Russian)]
Baubec, T., Dinh, H.Q., Pecinka, A., et al., Cooperation of multiple chromatin modifications can generate unanticipated stability of epigenetic states in Arabidopsis, Plant Cell, 2010, vol. 22, pp. 34–47.
Berger, S.L., The complex language of chromatin regulation during transcription, Nature, 2007, vol. 447, pp. 407–412.
Bernatavichute, Y.V., Zhang, X., Cokus, S., et al., Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana, PLoS One, 2008, vol. 3, art. e3156.
Birnbaum, K.D. and Roudier, F., Epigenetic memory and cell fate reprogramming in plants, Regeneration, 2016, vol. 4, pp. 15–20.
Blein, T., Pulido, A., Vialette-Guiraud, A., et al., A conserved molecular framework for compound leaf development, Science, 2008, vol. 322, no. 5909, pp. 1835–1839.
Bloomer, R.H., Hutchison, C.E., Bäurle, I., et al., The Arabidopsis epigenetic regulator ICU11 as an accessory protein of Polycomb Repressive Complex 2, Proc. Natl. Acad. Sci. U. S. A., 2020, vol. 117, no. 28, pp. 16660–16666.
Bondada, R., Somasundaram, S., Marimuthu, M.P., et al., Natural epialleles of Arabidopsis SUPERMAN display superwoman phenotypes, Commun. Biol., 2020, vol. 3, no. 1, p. 772.
Bowman, J.L., Sakai, H., Jack, T., et al., SUPERMAN, a regulator of floral homeotic genes in Arabidopsis, Development, 1992, vol. 114, pp. 599–615.
Bratzel, F., López-Torrejón, G., Koch, M., et al., Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination, Curr. Biol., 2010, vol. 26, no. 20, pp. 1853–1859.
Bräutigam, K. and Cronk, Q., DNA methylation and the evolution of developmental complexity in plants, Front. Plant Sci., 2018, vol. 9, p. 1447.
Catoni, M., Griffiths, J., Becker, C., et al., DNA sequence properties that predict susceptibility to epiallelic switching, EMBO J., 2017, vol. 36, pp. 617–628.
Charbonnel, C., Niazi, A.K., Elvira-Matelot, E., et al., The siRNA suppressor RTL1 is redox-regulated through glutathionylation of a conserved cysteine in the double-stranded-RNA-binding domain, Nucleic Acids Res., 2017, vol. 45, pp. 11891–11907.
Chen, D., Molito, A., Liu, C., et al., The Arabidopsis PRC1-like ring-finger proteins are necessary for repression of embryonic traits during vegetative growth, Cell Res., 2010, vol. 20, pp. 1332–1344.
Chen, X., Ding, A.B., and Zhong, X., Functions and mechanisms of plant histone deacetylases, Sci. China Life Sci., 2020, vol. 63, pp. 206–216.
Chen, J., Liu, J., Jiang, J., et al., F-box protein CFK1 interacts with and degrades de novo DNA methyltransferase in Arabidopsis, New Phytol., 2021, vol. 229, no. 6, pp. 3303–3317.
Colicchio, J., Kelly, J., and Hileman, L., Mimulus sRNAs are wound responsive and associated with transgenerationally plastic genes but rarely both, Int. J. Mol. Sci., 2020, vol. 21, no. 20, p. 7552.
Couturier, J., Chibani, K., Jacquot, J.P., and Rouhier, N., Cysteine-based redox regulation and signaling in plants, Front. Plant Sci., 2013, vol. 4, p. 105.
Cubas, P., Vincent, C., and Coen, E., An epigenetic mutation responsible for natural variation in floral symmetry, Nature, 1999, vol. 401, pp. 157–161.
Derkacheva, M. and Hennig, L., Variations on a theme: Polycomb group proteins in plants, J. Exp. Bot., 2014, vol. 65, no. 10, pp. 2769–2784.
Du, J., Johnson, L.M., Jacobsen, S.E., and Patel, D.J., DNA methylation pathways and their crosstalk with histone methylation, Nat. Rev. Mol. Cell Biol., 2015, vol. 16, pp. 519–532.
Duan, C.G., Wang, X.G., Tang, K., et al., MET18 connects the cytosolic iron-sulfur cluster assembly pathway to active DNA demethylation in Arabidopsis, PLoS Genet., 2015, vol. 11, art. e1005559.
Duan, C.G., Wang, X., Xie, S., et al., A pair of transposon-derived proteins function in a histone acetyltransferase complex for active DNA demethylation, Cell Res., 2016, vol. 27, no. 2, pp. 226–240.
Duan, C.G., Wang, X., Zhang, L., et al., A protein complex regulates RNA processing of intronic heterochromatin-containing genes in Arabidopsis, Proc. Natl. Acad. Sci. U. S. A., 2017, vol. 114, no. 35, pp. E7377–E7384.
Duempelmann, L., Skribbe, M., and Bühler, M., Small RNAs in the transgenerational inheritance of epigenetic information, Trends Genet., 2020, vol. 36, pp. 203–214.
Erdmann, R.M. and Picard, C.L., RNA-directed DNA methylation, PLoS Genet., 2020, vol. 16, no. 10, art. e1009034.
Ezhova, T.A. and Kupriyanova, E.V., Studying auxin’s role in ectopic outgrowths’ development on leaves of the Arabidopsis thaliana taeniata mutant, Russ. J. Dev. Biol., 2019, vol. 50, pp. 243–249.
Fal, K., Tomkova, D., Vachon, G., et al., Chromatin manipulation and editing: challenges, new technologies and their use in plants, Int. J. Mol. Sci., 2021, vol. 22, no. 2, p. 512.
Fedotov, A.P., Ezhova, T.A., and Timonin, A.C., Bizarre lamina margins in tae mutant of Arabidopsis thaliana (L.) Heynh. (Brassicaceae), Wulfenia, 2017, vol. 24, pp. 163–170.
Fenley, A.T., Anandakrishnan, R., Kidane, Y.H., and Onufriev, A.V., Modulation of nucleosomal DNA accessibility via charge-altering post-translational modifications in histone core, Epigenet. Chromatin, 2018, vol. 11, no. 1, p. 11.
Fletcher, J.C., State of the art: trxG factor regulation of post-embryonic plant development, Front. Plant Sci., 2017, vol. 8, p. 1925.
Förderer, A., Zhou, Y., and Turck, F., The age of multiplexity: recruitment and interactions of Polycomb complexes in plants, Curr. Opin. Plant Biol., 2016, vol. 29, pp. 169–178.
Foyer, C.H. and Noctor, G., Stress-triggered redox signalling: what’s in prospect?, Plant, Cell Environ., 2016, vol. 39, pp. 951–964.
Fritz, M.A., Rosa, S., and Sicard, A., Mechanisms underlying the environmentally induced plasticity of leaf morphology, Front. Genet., 2018, vol. 9, p. 478.
Fultz, D., Choudury, S.G., and Slotkin, R.K., Silencing of active transposable elements in plants, Curr. Opin. Plant Biol., 2015, vol. 27, pp. 67–76.
Gaillochet, C. and Lohmann, J.U., The never-ending story: from pluripotency to plant developmental plasticity, Development, 2015, vol. 142, no. 13, pp. 2237–2249.
Gallego-Bartolomé, J., DNA methylation in plants: mechanisms and tools for targeted manipulation, New Phytol., 2020, vol. 227, pp. 38–44.
Gallego-Bartolomé, J., Liu, W., Kuo, P.H., et al., Co-targeting RNA polymerases IV and V promotes efficient de novo DNA methylation in Arabidopsis, Cell, 2019, vol. 176, no. 5, pp. 1068–1082.
Le Goff, A., Allard, P., and Landecker, H., Heritable changeability: epimutation and the legacy of negative definition in epigenetic concepts, Stud. Hist. Philos. Sci., 2021, vol. 86, pp. 35–46.
Gouil, Q. and Baulcombe, D.C., DNA methylation signatures of the plant chromomethyltransferases, PLoS Genet., 2016, vol. 12, no. 12, р. e1006526.
Grimanelli, D. and Ingouff, M., DNA methylation readers in plants, J. Mol. Biol., 2020, vol. 432, pp. 1706–1717.
Guo, X., Xie, Q., Li, B., and Su, H., Molecular characterization and transcription analysis of DNA methyltransferase genes in tomato (Solanum lycopersicum), Genet. Mol. Biol., 2020, vol. 43, no. 1, р. e20180295.
Hallé, F., Modular growth in seed plants, Philos. Trans. R. Soc. Lond. B, 1986, vol. 313, pp. 77–88.
Handa, V. and Jeltsch, A., Profound flanking sequence preference of Dnmt3a and Dnmt3b mammalian DNA methyltransferases shape the human epigenome, J. Mol. Biol., 2005, vol. 348, pp. 1103–1112.
Harris, C.J., Scheibe, M., Wongpalee, S.P., et al., A DNA methylation reader complex that enhances gene transcription, Science, 2018, vol. 362, pp. 1182–1186.
Harvey, Z.H., Chen, Y., and Jarosz, D.F., Protein-based inheritance: epigenetics beyond the chromosome, Mol. Cell, 2018, vol. 69, pp. 195–202.
Hasegawa, J., Sakamoto, T., Fujimoto, S., et al., Auxin decreases chromatin accessibility through the TIR1/AFBs auxin signaling pathway in proliferative cells, Sci. Rep., 2018, vol. 8, p. 7773.
Hauser, M.T., Aufsatz, W., Jonak, C., and Luschnig, C., Transgenerational epigenetic inheritance in plants, Biochim. Biophys. Acta, 2011, vol. 1809, pp. 459–468.
He, Y. and Li, Z., Epigenetic environmental memories in plants: establishment, maintenance, and reprogramming, Trends Genet., 2018, vol. 34, pp. 856–866.
Heard, E. and Martienssen, R.A., Transgenerational epigenetic inheritance: myths and mechanisms, Cell, 2014, vol. 157, pp. 95–109.
Henderson, I.R. and Jacobsen, S.E., Epigenetic inheritance in plants, Nature, 2007, vol. 447, pp. 418–424.
Herms, D.A. and Mattson, W.J., The dilemma of plants: to grow or defend, Q. Rev. Biol., 1992, vol. 67, pp. 283–335.
Hofmeister, B.T., Lee, K., Rohr, N.A., et al., Stable inheritance of DNA methylation allows creation of epigenotype maps and the study of epiallele inheritance patterns in the absence of genetic variation, Genome Biol., 2017, vol. 18, no. 1, p. 155.
Huot, B., Yao, J., Montgomery, B.L., and He, S.Y., Growth-defense tradeoffs in plants: a balancing act to optimize fitness, Mol. Plant, 2014, vol. 7, no. 8, pp. 1267–1287.
Irish, V.F., Cell lineage in plant development, Curr. Opin. Cell Biol., 1991, vol. 3, pp. 983–987.
Jablonka, E. and Raz, G., Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution, Q. Rev. Biol., 2009, vol. 84, pp. 131–176.
Jacobsen, S.E. and Meyerowitz, E.M., Hypermethylated SUPERMAN epigenetic alleles in Arabidopsis, Science, 1997, vol. 277, no. 5329, pp. 1100–1103.
Jmii, S. and Cappadocia, L., Plant SUMO E3 ligases: function, structural organization, and connection with DNA, Front. Plant Sci., 2021, vol. 12, p. 652170.
Johannes, F. and Schmitz, R.J., Spontaneous epimutations in plants, New Phytol., 2019, vol. 221, pp. 1253–1259.
Kakutani, T., Epi-alleles in plants: inheritance of epigenetic information over generations, Plant Cell Physiol., 2002, vol. 43, no. 10, pp. 1106–1111.
Kalinkina, V.A., Onthogenetic polyvariaty in the representatives of Trifolium L. genus Lupinaster (Fabr.) Ser. section, Russ. J. Dev. Biol., 2017, vol. 48, no. 2, pp. 150–158.
Kim, D.H., Xi, Y., and Sung, S., Modular function of long noncoding RNA, COLDAIR, in the vernalization response, PLoS Genet., 2017, vol. 13, pp. 1–18.
Kim, J., Joo, Y., Kyung, J., et al., A molecular basis behind heterophylly in an amphibious plant, PLoS Genet., 2018, vol. 14, no. 2, art. e1007208.
Kishimoto, N., Sakai, H., Jackson, J., et al., Site specificity of the Arabidopsis METI DNA methyltransferase demonstrated through hypermethylation of the superman locus, Plant. Mol. Biol., 2001, vol. 46, no. 2, pp. 171–183.
Kol’tsov, N.K., Lotsi’s views on the evolution of organisms, Priroda (Moscow, Russ. Fed.), 1915, no. 10, p. 1253.
Kong, X., Hong, Y., Hsu, Yi-F., et al., SIZ1-mediated sumoylation of ROS1 enhances its stability and positively regulates active DNA demethylation in Arabidopsis, Mol. Plant, 2020, vol. 13, no. 12, pp. 1816–1824.
Kouzarides, T., Chromatin modifications and their function, Cell, 2007, vol. 128, pp. 693–705.
Kribelbauer, J.F., Lu, X.J., Rohs, R., et al., Toward a mechanistic understanding of DNA methylation readout by transcription factors, J. Mol. Biol., 2019, vol. 432, no. 6, pp. 1801–1815.
Kupriyanova, E.V., Denisov, E.R., Baier, M.A., and Ezhova, T.A., Differences in the manifestation of cell pluripotence in vivo and in vitro in the mutant Arabidopsis thaliana with the phenotype of cell memory disorder, Russ. J. Plant Physiol., 2021, vol. 68, no. 1, pp. 46–55.
Lämke, J. and Bäurle, I., Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants, Genome Biol., 2017, vol. 18, no. 1, p. 124.
Lardon, R. and Geelen, D., Natural variation in plant pluripotency and regeneration, Plants (Basel), 2020, vol. 9, no. 10, p. 1261.
Law, J.A. and Jacobsen, S.E., Establishing, maintaining and modifying DNA methylation patterns in plants and animals, Nat. Rev. Genet., 2010, vol. 11, pp. 204–220.
Le, N.T., Harukawa, Y., Miura, S., et al., Epigenetic regulation of spurious transcription initiation in Arabidopsis, Nat. Commun., 2020, vol. 11, p. 3224.
Li, S.W., Molecular bases for the regulation of adventitious root generation in plants, Front. Plant Sci., 2021, vol. 12, p. 614072.
Li, Y. and Li, H., Many keys to push: diversifying the ‘readership’ of plant homeodomain fingers, Acta Biochim. Biophys. Sin. (Shanghai), 2012, vol. 44, no. 1, pp. 28–39.
Li, B., Carey, M., and Workman, J.L., The role of chromatin during transcription, Cell, 2007, vol. 128, pp. 707–719.
Li, G., Hu, S., Yang, J., et al., Water-wisteria as an ideal plant to study heterophylly in higher aquatic plants, Plant Cell Rep., 2017a, vol. 36, pp. 1225–1236.
Li, G., Hu, S., Yang, J., et al., Water-wisteria as an ideal plant to study heterophylly in higher aquatic plants, Plant Cell Rep., 2017b, vol. 36, pp. 1225–1236.
Li, X., Harris, C.J., Zhong, Z., et al., Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation, Proc. Natl. Acad. Sci. U. S. A., 2018a, vol. 115, pp. E8793–E8802.
Li, Y., Kumar, S., and Qian, W., Active DNA demethylation: mechanism and role in plant development, Plant Cell Rep., 2018b, vol. 37, no. 1, pp. 77–85.
Li, J., Yang, D.-L., Huang, H., et al., Epigenetic memory marks determine epiallele stability at loci targeted by de novo DNA methylation, Nat. Plants, 2020, vol. 6, pp. 661–674.
Liang, S.C., Hartwig, B., Perera, P., et al., Kicking against the PRCs—a domesticated transposase antagonises silencing mediated by Polycomb group proteins and is an accessory component of Polycomb repressive complex 2, PLoS Genet., 2015, vol. 11, no. 12, art. e1005660.
Liégard, B., Baillet, V., Etcheverry, M., et al., Quantitative resistance to clubroot infection mediated by transgenerational epigenetic variation in Arabidopsis, New Phytol., 2019, vol. 222, pp. 468–479.
Lindermayr, C., Rudolf, E.E., Durner, J., and Groth, M., Interactions between metabolism and chromatin in plant models, Mol. Metab., 2020, vol. 38, p. 100951.
Lisch, D., How important are transposons for plant evolution?, Nat. Rev. Genet., 2013, vol. 14, pp. 49–61.
Liu, C., Lu, F., Cui, X., and Cao, X., Histone methylation in higher plants, Annu. Rev. Plant Biol., 2010, vol. 61, pp. 395–420.
Liu, R. and Lang, Z., The mechanism and function of active DNA demethylation in plants, J. Integr. Plant Biol., 2019, vol. 62, pp. 148–159.
Locato, V., Cimini, S., and De Gara, L., ROS and redox balance as multifaceted players of cross-tolerance: epigenetic and retrograde control of gene expression, J. Exp. Bot., 2018, vol. 69, pp. 3373–3391.
Lodha, M., Marco, C.F., and Timmermans, M.C., Genetic and epigenetic regulation of stem cell homeostasis in plants, Cold Spring Harb. Symp. Quant. Biol., 2008, vol. 73, pp. 243–251.
Ma, X., Lv, S., Zhang, C., et al., Histone deacetylases and their functions in plants, Plant Cell Rep., 2013, vol. 32, pp. 465–478.
Manning, K., Tör, M., Poole, M., et al., A naturally occurring epigenetic mutation in a gene encoding an sbp-box transcription factor inhibits tomato fruit ripening, Nat. Genet., 2006, vol. 38, no. 8, pp. 948–952.
Mateo-Bonmati, E., Esteve-Bruna, D., Juan-Vicente, L., et al, INCURVATA11 and CUPULIFORMIS2 are redundant genes that encode epigenetic machinery components in Arabidopsis, Plant Cell, 2018, vol. 30, pp. 1596–1616.
Matzke, M.A. and Mosher, R.A., RNA-directed DNA methylation: an epigenetic pathway of increasing complexity, Nat. Rev. Genet., 2014, vol. 15, pp. 394–408.
Miura, K., Agetsuma, M., Kitano, H., et al., A metastable dwarf1 epigenetic mutant affecting plant stature in rice, Proc. Natl. Acad. Sci. U. S. A., 2009, vol. 106, no. 27, pp. 11218–11223.
Morange, M., The attempt of Nikolai Koltzoff (Koltsov) to link genetics, embryology and physical chemistry, J. Biosci., 2011, vol. 36, no. 211–214.
Morgado, L., Preite, V., Oplaat, C., et al., Small RNAs reflect grandparental environments in apomictic dandelion, Mol. Biol. Evol., 2017, vol. 34, pp. 2035–2040.
Mozgova, I. and Hennig, L., The Polycomb group protein regulatory network, Annu. Rev. Plant Biol., 2015, vol. 66, pp. 269–296.
Münzbergová, Z., Latzel, V., Šurinová, M., and Hadincová, V., DNA methylation as a possible mechanism affecting ability of natural populations to adapt to changing climate, Oikos, 2019, vol. 128, no. 1, pp. 124–134.
Muyle, A., Shearn, R., and Marais, G.A., The evolution of sex chromosomes and dosage compensation in plants, Genome Biol. Evol., 2017, vol. 9, no. 3, pp. 627–645.
Muyle, A., Zemp, N., Fruchard, C., et al., Genomic imprinting mediates dosage compensation in a young plant XY system, Nat. Plants, 2018, vol. 4, no. 9, pp. 677–680.
Nakayama, H., Nakayama, N., Seiki, S., et al., Regulation of the knox-ga gene module induces heterophyllic alteration in North American lake cress, Plant Cell, 2014, vol. 26, pp. 4733–4748.
Narlikar, G.J., Sundaramoorthy, R., and Owen-Hughes, T., Mechanisms and functions of atp-dependent chromatin-remodeling enzymes, Cell, 2013, vol. 154, pp. 490–503.
Nashun, B., Hill, P.W., and Hajkova, P., Reprogramming of cell fate: epigenetic memory and the erasure of memories past, EMBO J., 2015, vol. 34, no. 10, pp. 1296–1308.
Noble, D., Conrad Waddington and the origin of epigenetics, J. Exp. Biol., 2015, vol. 218, pp. 816–818.
Notov, A.A. and Zhukova, L.A., The concept of ontogenesis polyvariance and modern evolutionary morphology, Biol. Bull. (Moscow), 2019, vol. 46, no. 1, pp. 47–55.
Oborny, B., The plant body as a network of semi-autonomous agents: a review, Philos. Trans. R. Soc. Lond., B, 2019, vol. 374, no. 1774, p. 20180371.
O’Malley, R.C. and Ecker, J.R., Epiallelic variation in Arabidopsis thaliana, Cold Spring Harb. Symp. Quant. Biol., 2012, vol. 77, pp. 135–145.
Ornelas-Ayala, D., Garay-Arroyo, A., Garcia-Ponce, B., et al., The epigenetic faces of ULTRAPETALA1, Front. Plant Sci., 2021, vol. 12, p. 637244.
Pagliarani, C. and Gambino, G., Small RNA mobility: spread of RNA silencing effectors and its effect on developmental processes and stress adaptation in plants, Int. J. Mol. Sci., 2019, vol. 20, no. 17, p. 4306.
Parrilla-Doblas, J.T., Roldan-Arjona, T., Ariza, R.R., and Cordoba-Canero, D., Active DNA demethylation in plants, Int. J. Mol. Sci., 2019, vol. 20, no. 19, p. 4683.
Patel, D.J., A structural perspective on readout of epigenetic histone and DNA methylation marks, Cold Spring Harb. Perspectives Biol., 2016, vol. 8, no. 3, art. a018754.
Pikaard, C.S. and Scheid, O.M., Epigenetic regulation in plants, Cold Spring Harb. Perspectives Biol., 2014, vol. 6, no. 12, art. a019315.
Poethig, S., Genetic mosaics and cell lineage analysis in plants, Trends Genet., 1989, vol. 5, pp. 273–277.
Prakash, S., Singh, R., and Lodhi, N., Histone demethylases and control of gene expression in plants, Cell. Mol. Biol., 2014, vol. 60, no. 5, pp. 97–105.
Ptashne, M., Epigenetics: core misconcept, Proc. Natl. Acad. Sci. U. S. A., 2013, vol. 110, pp. 7101–7103.
Raju, S.K.K., Ritter, E.J., and Niederhuth, C.E., Establishment, maintenance, and biological roles of non-CG methylation in plants, Essays Biochem., 2019, vol. 63, pp. 743–755.
Ramenskii, E.V., Epigenetics: Waddington or Koltsov?, Ontogenez., 2018, vol. 49, no. 6, pp. 391–396.
Razin, S.V. and Bystritskii, A.A., Khromatin: upakovannyi genom (Chromatin: Packed Genome), Moscow: Binom. Laboratoriya znanii, 2009.
Rendina González, A.P., Preite, V., Verhoeven, K.J.F., and Latzel, V., Transgenerational effects and epigenetic memory in the clonal plant Trifolium repens, Front. Plant Sci., 2018, vol. 9, p. 1677.
Rohde, A., Grunau, C., De Beck, L., et al., Carpel, a new Arabidopsis epi-mutant of the SUPERMAN gene: phenotypic analysis and DNA methylation status, Plant Cell Physiol., 1999, vol. 40, no. 9, pp. 961–972.
Rohini, G., Romika, K., Sneha, T., and Shweta, G., Genomic survey, gene expression analysis and structural modeling suggest diverse roles of DNA methyltransferases in legumes, PLoS One, 2014, vol. 2, art. e88947.
Roudier, F., Ahmed, I., Berard, C., et al., Integrative epigenomic mapping defines four main chromatin states in Arabidopsis, EMBO J., 2011, vol. 30, pp. 1928–1938.
Saravana Kumar, R.M., Wang, Y., Zhang, X., et al., Redox components: key regulators of epigenetic modifications in plants, Int. J. Mol. Sci., 2020, vol. 21, no. 4, p. 1419.
Satterlee, J.W. and Scanlon, M.J., Coordination of leaf development across developmental axes, Plants (Basel), 2019, vol. 8, no. 10, p. 433.
Scheid, R., Chen, J., and Zhong, X., Biological role and mechanism of chromatin readers in plant, Curr. Opin. Plant Biol., 2021, vol. 10, no. 61, p. 102008.
Schmitz, R.J., Schultz, M.D., Lewsey, M.G., et al., Transgenerational epigenetic instability is a source of novel methylation variants, Science, 2011, vol. 334, no. 6054, pp. 369–373.
Selma, S. and Orzáez, D., Perspectives for epigenetic editing in crops, Transgenic Res., 2021.
Serebryakov, I.G. and Serebryakova, T.I., Some problems of the evolution of life forms of flowering plants, Bot. Zh., 1972, vol. 57, no. 5, pp. 417–433.
Seta, A., Tabara, M., Nishibori, Y., et al., Post-translational regulation of the dicing activities of Arabidopsis DICER-LIKE 3 and 4 by inorganic phosphate and the redox state, Plant Cell Physiol., 2017, vol. 58, pp. 485–495.
Shi, W., Chen, X., Gao, L., et al., Transient stability of epigenetic population differentiation in a clonal invader, Front. Plant Sci., 2019, vol. 9, p. 1851.
Shin, J., Bae, S., and Seo, P.J., De novo shoot organogenesis during plant regeneration, J. Exp. Bot., 2020, vol. 71, no. 1, pp. 63–72. https://doi.org/10.1093/jxb/erz395
Sijacic, P., Holder, D.H., Bajic, M., and Deal, R.B., Deal R.B. Methyl-CpG-binding domain 9 (MBD9) is required for H2A.Z incorporation into chromatin at a subset of H2A.Z-enriched regions in the Arabidopsis genome, PLoS Genet., 2019, vol. 15, no. 8, p. 1008326.
Song, Q. and Chen, Z.J., Epigenetic and developmental regulation in plant polyploids, Curr. Opin. Plant Biol., 2015, vol. 24, pp. 101–109.
Stoddard, C.I., Feng, S., Campbell, M.G., et al., A nucleosome bridging mechanism for activation of a maintenance DNA methyltransferase, Mol. Cell, 2019, vol. 73, pp. 73–83.
Sultan, S.E., Developmental plasticity: re-conceiving the genotype, Interface Focus, 2017, vol. 7, no. 5, p. 20170009.
Surguchov, A., Emamzadeh, F.N. and Surguchev, A.A., Amyloidosis and longevity: a lesson from plants, Biology, 2019, vol. 8, no. 2, p. 43.
Szymkowiak, E.J. and Sussex, I.M., What chimeras can tell us about plant development, Annu. Rev. Plant Physiol. Plant Mol. Biol., 1996, vol. 47, pp. 351–376.
Tamiru, M., Hardcastle, T.J., and Lewsey, M.G., Regulation of genome-wide DNA methylation by mobile small RNAs, New Phytol., 2018, vol. 217, pp. 540–546.
Tikhodeev, O.N., Epigenetic and eugenetic processes, Usp. Sovrem. Biol., 2015, vol. 135, no. 6, pp. 542–553.
Torres, I.O. and Fujimori, D.G., Functional coupling between writers, erasers and readers of histone and DNA methylation, Curr. Opin. Struct. Biol., 2015, vol. 35, pp. 68–75.
Tricker, P.J. and Gibbings, J.G., Rodríguez, et al., Low relative humidity triggers RNA-directed de novo DNA methylation and suppression of genes controlling stomatal development, J. Exp. Bot., 2012, vol. 63, pp. 3799–3813.
Tricker, P.J., López, C.M., Gibbings, G., et al., Transgenerational, dynamic methylation of stomata genes in response to low relative humidity, Int. J. Mol. Sci., 2013, vol. 14, pp. 6674–6689.
Ullah, F., Hamilton, M., Reddy, A.S.N., and Ben-Hur, A., Exploring the relationship between intron retention and chromatin accessibility in plants, BMC Genomics, 2018, vol. 19, p. 21.
Vanyushin, B.F., DNA methylation and epigenetics, Russ. J. Genet., 2006, vol. 42, no. 9, pp. 985–997.
Velanis, C.N., Perera, P., Thomson, B., et al., The domesticated transposase alp2 mediates formation of a novel Polycomb protein complex by direct interaction with msi1, a core subunit of Polycomb repressive complex 2 (PRC2), PLoS Genet., 2020, vol. 16, no. 5, art. e1008681.
Waddington, C.H., The basic ideas of biology, in Towards a Theoretical Biology, Waddington, C.H., Ed., Edinburgh: Edinburgh University Press, 1968, vol. 1, pp. 1–32.
Wang, M., Zhang, K., Ngo, V., et al., Identification of DNA motifs that regulate DNA methylation, Nucleic Acids Res., 2019, vol. 47, no. 13, pp. 6753–6768.
Wei, G., Liu, K., Shen, T., et al., Position-specific intron retention is mediated by the histone methyltransferase SDG725, BMC Biol., 2018, vol. 16, p. 44.
Weigel, D. and Colot, V., Epialleles in plant evolution, Genome Biol., 2012, vol. 13, no. 10, p. 249.
Whipple, C., Defining the plant germ line-nature or nurture?, Science, 2012, vol. 337, no. 6092, pp. 301–302.
Wienholz, B.L., Kareta, M.S., Moarefi, A.H., et al., DNMT3L modulates significant and distinct flanking sequence preference for DNA methylation by DNMT3A and DNMT3B in vivo, PLoS Genet., 2010, vol. 6, art. e1001106.
Williams, B.P. and Gehring, M., Principles of epigenetic homeostasis shared between flowering plants and mammals, Trends Genet., 2020, vol. 36, no. 10, pp. 751–763.
Wu, M.F., Yamaguchi, N., Xiao, J., et al., Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate, Elife, 2015, vol. 4, art. e09269.
Wu, H.W., Deng, S., Xu, H., et al., A noncoding RNA transcribed from the AGAMOUS (AG) second intron binds to CURLY LEAF and represses AG expression in leaves, New Phytol., 2018, vol. 219, pp. 1480–1491.
Wu, Z., Fang, X., Zhu, D., and Dean, C., Autonomous pathway: FLOWERING LOCUS C repression through an antisense-mediated chromatin-silencing mechanism, Plant Physiol., 2020, vol. 182, pp. 27–37.
Xiao, J. and Wagner, D., Polycomb repression in the regulation of growth and development in Arabidopsis, Curr. Opin. Plant Biol., 2015, vol. 23, pp. 15–24.
Xiao, X., Zhang, J., Li, T., et al., A group of SUVH methyl-DNA binding proteins regulate expression of the DNA demethylase ROS1 in Arabidopsis, J. Integr. Plant Biol., 2019, vol. 61, no. 2, pp. 110–119.
Xu, L., De novo root regeneration from leaf explants: wounding, auxin, and cell fate transition, Curr. Opin. Plant Biol., 2018, vol. 41, pp. 39–45.
Xu, Y., Zhang, L., and Wu, G., Epigenetic regulation of juvenile-to-adult transition in plants, Front. Plant Sci., 2018a, vol. 9, p. 1048.
Xu, F., Feng, X., Chen, X., et al., Cytoplasmic argonaute protein promotes the inheritance of RNAi, Cell Rep., 2018b, vol. 23, pp. 2482– 2494.
Yan, B., Lv, Y., Zhao, C., and Wang, X., Knowing when to silence: roles of Polycomb-group proteins in SAM maintenance, root development, and developmental phase transition, Int. J. Mol. Sci., 2020, vol. 21, no. 16, p. 5871.
Yu, Z., Zhang, G., Teixeira da Silva, J.A., et al., DNA methyltransferase and demethylase gene families in Dendrobium officinale reveal their potential functions in polysaccharide accumulation, BMC Plant Biol., 2021, vol. 21, p. 21.
Zhang, X., Clarenz, O., Cokus, S., et al., Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis, PLoS Biol., 2007, vol. 5, art. e129.
Zhang, X., Bernatavichute, Y.V., Cokus, S., et al., Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana, Genome Biol., 2009, vol. 10, no. 6, p. R62.
Zhang, L., Cheng, Z., Qin, R., et al., Identification and characterization of an epi-allele of FIE1 reveals a regulatory linkage between two epigenetic marks in rice, Plant Cell, 2012, vol. 24, no. 11, pp. 4407–4421.
Zhang, H.M., Lang, Z.B., and Zhu, J.K., Dynamics and function of DNA methylation in plants, Nat. Rev. Mol. Cell Biol., 2018, vol. 19, pp. 489–506.
Zhang, Y.Z., Lin, J., Ren, Z., et al., Genome-wide distribution and functions of the AAE complex in epigenetic regulation in Arabidopsis, J. Integr. Plant Biol., 2021, vol. 63, no. 4, pp. 707–722.
Zheng, X., Chen, L., Xia, H., et al., Transgenerational epimutations induced by multi-generation drought imposition mediate rice plant’s adaptation to drought condition, Sci. Rep., 2017, vol. 7, p. 39843.
Zhu, H., Wang, G., and Qian, J., Transcription factors as readers and effectors of DNA methylation, Nat. Rev. Genet., 2016, vol. 17, pp. 551–565.
Zhukova, L.A. and Glotov, N.V., Morphological polyvariance of ontogeny in natural plant populations, Russ. J. Dev. Biol., 2001, vol. 32, no. 6, pp. 381–387.
Funding
The work was performed with the financial support of a grant of the Russian Foundation for Basic Research (project nos. 19-04-00149 and 20-14-50020).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The author declares that she has no conflicts of interest. This article does not contain any studies involving human participants or laboratory animals as experimental models performed by the authors.
Additional information
Translated by A. Ermakov
Abbreviations: EM—epigenetic modifications; ER—epigenetic regulators; 5mC—5-methylcytosine; TF—transcription factor; DMR—differentially methylated regions of the genome.
Supplementary Information
The online version contains supplementary material available at https://doi.org/10.1134/S1062360421060047.
Rights and permissions
Open Access. This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ezhova, T.A. Paradoxes of Plant Epigenetics. Russ J Dev Biol 52, 355–369 (2021). https://doi.org/10.1134/S1062360421060047
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1062360421060047